A CRISPR approach to gene targeting - PubMed (original) (raw)
A CRISPR approach to gene targeting
Dana Carroll. Mol Ther. 2012 Sep.
No abstract available
Figures
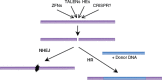
Figure 1
Consequences of targeted genomic cleavage. A double-strand break made by any type of cleavage reagent can be repaired by error-prone nonhomologous end joining (NHEJ), leaving small insertions and/or deletions at the site. An alternative mode of repair is homologous recombination (HR), which can use a manipulated donor DNA as a template, resulting in replacement of genomic sequences. The break can be made by any targetable nuclease: zinc-finger nucleases (ZFNs), transcription activator–like effector nucleases (TALENs), homing endonucleases (HEs), or, perhaps, the new CRISPR reagents.
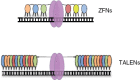
Figure 2
ZFNs and TALENs. (Top) Each zinc finger (small ovals) in a zinc-finger nuclease (ZFN) binds primarily to three consecutive base pairs; a minimum of three fingers is required to provide sufficient affinity. Different colors indicate fingers recognizing different DNA triplets. Each set of fingers is joined to a _Fok_I-derived cleavage domain (large ovals) by a short linker. (Bottom) In transcription activator–like effector nucleases (TALENs), each module (small ovals) binds a single base pair; the four colors indicate modules for each of the four base pairs. The minimum effective number of modules is 10–12, but more are typically used. The linker to the _Fok_I domain (large ovals) is longer than for ZFNs and contains additional TALE-derived sequences.
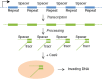
Figure 3
The type II CRISPR system. In a bacterial genome, identical repeats flank virus- or plasmid-derived spacer sequences in tandem arrays (blue). Long transcripts (green line) are processed into short RNAs containing a single spacer and a partial repeat. These short RNAs form partial duplexes with tracrRNAs and are bound by the Cas9 protein (orange oval). The complex then cleaves invading viral or plasmid DNA directed by the spacer RNAs. tracrRNA, _trans_-activated CRISPR RNA.
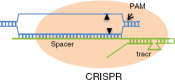
Figure 4
The CRISPR minimal-cleavage elements described by Jinek et al. A single RNA (green lines) with the critical elements of spacer and tracrRNA binds Cas9 protein (orange oval) and directs cleavage (arrowheads) to a sequence in DNA (blue) that has homology to the spacer. The region of RNA–DNA base pairing provides cleavage specificity. The target must also have a particular two– to three–base pair sequence adjacent to the region of homology, called PAM, which is recognized by the complex. PAM, protospacer adjacent motif; tracrRNA, _trans_-activated CRISPR RNA.
Similar articles
- Gene targeting in Drosophila and Caenorhabditis elegans with zinc-finger nucleases.
Carroll D, Beumer KJ, Morton JJ, Bozas A, Trautman JK. Carroll D, et al. Methods Mol Biol. 2008;435:63-77. doi: 10.1007/978-1-59745-232-8_5. Methods Mol Biol. 2008. PMID: 18370068 - Gene targeting using zinc finger nucleases.
Porteus MH, Carroll D. Porteus MH, et al. Nat Biotechnol. 2005 Aug;23(8):967-73. doi: 10.1038/nbt1125. Nat Biotechnol. 2005. PMID: 16082368 Review. - Beyond TALENs.
Pennisi E. Pennisi E. Science. 2012 Dec 14;338(6113):1411. doi: 10.1126/science.338.6113.1411. Science. 2012. PMID: 23239710 No abstract available. - The tale of the TALEs.
Pennisi E. Pennisi E. Science. 2012 Dec 14;338(6113):1408-11. doi: 10.1126/science.338.6113.1408. Science. 2012. PMID: 23239709 No abstract available. - The CRISPR/Cas9 system for plant genome editing and beyond.
Bortesi L, Fischer R. Bortesi L, et al. Biotechnol Adv. 2015 Jan-Feb;33(1):41-52. doi: 10.1016/j.biotechadv.2014.12.006. Epub 2014 Dec 20. Biotechnol Adv. 2015. PMID: 25536441 Review.
Cited by
- Integrative Comparison of the Role of the PHOSPHATE RESPONSE1 Subfamily in Phosphate Signaling and Homeostasis in Rice.
Guo M, Ruan W, Li C, Huang F, Zeng M, Liu Y, Yu Y, Ding X, Wu Y, Wu Z, Mao C, Yi K, Wu P, Mo X. Guo M, et al. Plant Physiol. 2015 Aug;168(4):1762-76. doi: 10.1104/pp.15.00736. Epub 2015 Jun 16. Plant Physiol. 2015. PMID: 26082401 Free PMC article. - The basic building blocks and evolution of CRISPR-CAS systems.
Makarova KS, Wolf YI, Koonin EV. Makarova KS, et al. Biochem Soc Trans. 2013 Dec;41(6):1392-400. doi: 10.1042/BST20130038. Biochem Soc Trans. 2013. PMID: 24256226 Free PMC article. Review. - Molecular Cytogenetics in the Era of Chromosomics and Cytogenomic Approaches.
Liehr T. Liehr T. Front Genet. 2021 Oct 13;12:720507. doi: 10.3389/fgene.2021.720507. eCollection 2021. Front Genet. 2021. PMID: 34721522 Free PMC article. Review. - APOE genotype and stress response - a mini review.
Dose J, Huebbe P, Nebel A, Rimbach G. Dose J, et al. Lipids Health Dis. 2016 Jul 25;15:121. doi: 10.1186/s12944-016-0288-2. Lipids Health Dis. 2016. PMID: 27457486 Free PMC article. Review. - Towards DNA-free CRISPR/Cas9 genome editing for sustainable oil palm improvement.
Masani MYA, Norfaezah J, Bahariah B, Fizree MPMA, Sulaiman WNSW, Shaharuddin NA, Rasid OA, Parveez GKA. Masani MYA, et al. 3 Biotech. 2024 Jun;14(6):166. doi: 10.1007/s13205-024-04010-w. Epub 2024 May 28. 3 Biotech. 2024. PMID: 38817736 Review.
References
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS., and, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. - PubMed
- Bogdanove AJ., and, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources