Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria - PubMed (original) (raw)
Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria
Giedrius Gasiunas et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Clustered, regularly interspaced, short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems provide adaptive immunity against viruses and plasmids in bacteria and archaea. The silencing of invading nucleic acids is executed by ribonucleoprotein complexes preloaded with small, interfering CRISPR RNAs (crRNAs) that act as guides for targeting and degradation of foreign nucleic acid. Here, we demonstrate that the Cas9-crRNA complex of the Streptococcus thermophilus CRISPR3/Cas system introduces in vitro a double-strand break at a specific site in DNA containing a sequence complementary to crRNA. DNA cleavage is executed by Cas9, which uses two distinct active sites, RuvC and HNH, to generate site-specific nicks on opposite DNA strands. Results demonstrate that the Cas9-crRNA complex functions as an RNA-guided endonuclease with RNA-directed target sequence recognition and protein-mediated DNA cleavage. These findings pave the way for engineering of universal programmable RNA-guided DNA endonucleases.
Conflict of interest statement
Conflict of interest statement: R.B. and P.H. are employees of DuPont Nutrition & Health and G.G., R.B., P.H., and V.S. are inventors on patent applications related to CRISPR.
Figures
Fig. 1.
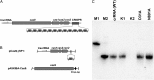
The Cas9 protein copurifies with crRNA. (A) Schematic representation of the CRISPR3/Cas system of S. thermophilus DGCC7710. Four cas genes (cas9, cas1, cas2, csn2) are located upstream of the CRISPR repeat-spacer array, consisting of 13 repeat (R) sequences and 12 unique spacers (S1–S12). The tracrRNA, required for crRNA maturation in type II CRISPR/Cas systems (21), is located upstream of the cas9 gene and is encoded on the opposite DNA strand (shown by an arrow) with respect to the other elements of this system. (B) Schematic representation of heterologous loci in two plasmids used for the coexpression of the Cas9–crRNA complex. E. coli RR1 contained pCas9(−)SP1 (encoding Cas1, Cas2, Csn2, SP1, and tracrRNA) and pASKIBA-Cas9 (encoding the Strep-tagged version of Cas9) plasmids. (C) Northern blot analysis of Cas9–crRNA complexes using anti-crDNA oligonucleotide as a probe. M1, 84-nt oligodeoxynucleotide corresponding to the spacer1-repeat unit; M2, 42-nt synthetic oligoribonucleotide corresponding to the predicted S. thermophilus CRISPR3 crRNA (
Fig. S2
); crRNA (WT), crRNA isolated from the WT Cas9–crRNA complex; K1, crRNA (WT) treated with DNase I for 15 min; K2, crRNA (WT) treated with RNase I for 15 min; D31A, crRNA purified from the Cas9 D31A mutant complex; N891A, crRNA purified from the Cas9 N891A mutant complex.
Fig. 2.
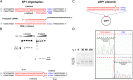
The Cas9–crRNA complex cleaves dsDNA within the protospacer. (A) Oligoduplex substrate used in the cleavage assay. The 55-nt oligoduplex SP1 contains the protospacer1 (red letters), PAM (blue letters), and 10-nt flanking sequences on both sides identical to those in pSP1 plasmid. In the SP1 oligoduplex, the DNA strand complementary to the 5′-terminal fragment of crRNA (red letters) is termed the “(+)strand,” and the opposite DNA strand is termed the “(−)strand.” (B) Oligoduplex SP1 cleavage. The Cas9–crRNA complex (2.5 M) and 1 nM SP1 oligoduplex labeled with 33P at the 5′ end of either the (+) or (–)strand were incubated in the reaction buffer [10 mM Tris⋅HCl (pH 7.5), 10 mM NaCl, 10 mM MgCl2, 0.1 mg/mL BSA] at 37 °C for various time intervals (30 s to 10 min), and reaction products were analyzed by 20% PAGE. Lanes M1 and M2 contain chemically synthesized 5′-end, 33P-labeled, 37-nt and 18-nt oligodeoxynucleotides corresponding to the cleavage products of (−) and (+) DNA strands, respectively. Cleavage positions are indicated by arrows. (C) Schematic representation of pSP1 plasmid (16) used in the plasmid cleavage assay. (D) pSP1 plasmid cleavage. (Left) Agarose gel analysis of pSP1 cleavage products. FLL, full-length linear DNA cut at both strands; OC, open circular DNA nicked at one of the strands; SC, supercoiled plasmid DNA. Final reaction mixtures at 37 °C contained 2.5 nM of the pSP1 plasmid and 2.5 nM of the Cas9–crRNA complex in the reaction buffer (see section B). (Right) Direct-sequencing electropherograms of (+) (Upper) and (–) (Lower) strands of the pSP1 plasmid cleavage product.
Fig. 3.
A PAM is required for in vitro DNA binding and cleavage by the Cas9–crRNA complex. (A) Agarose gel analysis of plasmid DNA cleavage products. Three different plasmids—PAM+Protospacer+ (pSP1 containing both the protospacer and the PAM), PAM+Protospacer− (pUC18 containing multiple PAMs but no protospacer), and PAM-Protospacer+ [pSP1-pΔ (16) containing a protospacer without a PAM]—were incubated at 2.5-nM concentration with 2 nM of the Cas9–crRNA complex in the reaction buffer [10 mM Tris⋅HCl (pH 7.5), 10 mM NaCl, 10 mM MgCl2, 0.1 mg/mL BSA] at 37 °C for various time intervals, and reaction products were analyzed by agarose gel electrophoresis. FLL, full- length linear DNA cut at both strands; OC, open circular DNA nicked at one of DNA strands; SC, supercoiled plasmid DNA. (B) Time courses of (+)strand hydrolysis in the single-stranded and double-stranded oligodeoxynucleotides. Reactions containing 2 nM Cas9–crRNA and 1 nM of oligodeoxynucleotide were conducted at 37 °C in the reaction buffer (see section A). SP1 (filled circles) and SP1-pΔ (open squares) oligoduplexes were used as dsDNA. s(+)SP1 (open triangles) and s(+)SP1-pΔ (filled squares) were used as ssDNA. (C and D) dsDNA and ssDNA (+)strand) binding by the Cas9–crRNA complex. The reactions contained 0.5 nM of the 33P-labeled ssDNA or dsDNA oligonucleotide and the protein at the concentrations indicated above each lane. After 15 min at room temperature, the samples were subjected to PAGE for 2 h and analyzed as described in Materials and Methods.
Fig. 4.
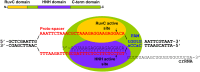
RuvC and HNH active-site motifs of Cas9 contribute to the cleavage of opposite DNA strands. (A) Localization of the conserved active-site motifs within Cas9 protein. Amino acid residues identified as crucial for Cas9 in vivo activity (16) are indicated. (B) Agarose gel analysis of pSP1 plasmid cleavage by Cas9 and mutant proteins. Reactions were performed as described in Materials and Methods. (C) Strand preference of the D31A mutant. Reactions were performed as described in Fig. 2_A_ and Materials and Methods. The D31 mutant cleaves only the (+)strand of the SP1 oligoduplex. (D) Strand preference of the N891A mutant. The N891 mutant cleaves only the (−)strand of the SP1 oligoduplex. Cleavage positions are indicated by arrows.
Fig. 5.
Schematic arrangement and mechanism of crRNA-directed DNA cleavage by the Cas9–crRNA complex. Domain architecture of Cas9 is shown schematically at the top. The Cas9–crRNA complex binds to the dsDNA containing a PAM. crRNA binds to the complementary (+)strand, resulting in DNA strand separation and R-loop formation. In the ternary complex, the RuvC active site of Cas9 is positioned at the scissile phosphate on the unpaired (−)strand, and the HNH active site is located at the scissile phosphate on the DNA (+)strand bound to crRNA. Coordinated action of both active sites results in the double-strand break 3 nt upstream the PAM, generating blunt-ended DNA.
Fig. P1.
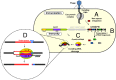
The Cas9–crRNA complex generates two distinct DNA nicks on opposing dsDNA strands that match the loaded small, interfering crRNA sequence. (A) After phage DNA entry into the cell, a piece of the invading DNA is inserted as a spacer into the CRISPR locus. (B) The CRISPR repeat-spacer array is transcribed and processed into short crRNAs. (C) The crRNA forms a ribonucleoprotein complex with Cas9, which recognizes invading DNA homologous to the crRNA sequence and mediates interference. (D) Invading DNA cleavage by Cas9–crRNA. In the presence of Mg2+ ions, the signature Cas9 protein nicks each DNA strand 3 nt upstream of the PAM sequence to generate blunt DNA ends through RuvC- and HNH-like active sites that act on separate DNA strands.
Similar articles
- crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus.
Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. Karvelis T, et al. RNA Biol. 2013 May;10(5):841-51. doi: 10.4161/rna.24203. Epub 2013 Mar 27. RNA Biol. 2013. PMID: 23535272 Free PMC article. - Programmable DNA cleavage in vitro by Cas9.
Karvelis T, Gasiunas G, Siksnys V. Karvelis T, et al. Biochem Soc Trans. 2013 Dec;41(6):1401-6. doi: 10.1042/BST20130164. Biochem Soc Trans. 2013. PMID: 24256227 Review. - The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA.
Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. Fonfara I, et al. Nature. 2016 Apr 28;532(7600):517-21. doi: 10.1038/nature17945. Epub 2016 Apr 20. Nature. 2016. PMID: 27096362 - A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. Jinek M, et al. Science. 2012 Aug 17;337(6096):816-21. doi: 10.1126/science.1225829. Epub 2012 Jun 28. Science. 2012. PMID: 22745249 Free PMC article. - DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes.
Plagens A, Richter H, Charpentier E, Randau L. Plagens A, et al. FEMS Microbiol Rev. 2015 May;39(3):442-63. doi: 10.1093/femsre/fuv019. Epub 2015 Apr 30. FEMS Microbiol Rev. 2015. PMID: 25934119 Free PMC article. Review.
Cited by
- The unsung heroes of CRISPR.
Ledford H. Ledford H. Nature. 2016 Jul 21;535(7612):342-4. doi: 10.1038/535342a. Nature. 2016. PMID: 27443723 No abstract available. - CRISPR-mediated gene modification of hematopoietic stem cells with beta-thalassemia IVS-1-110 mutation.
Gabr H, El Ghamrawy MK, Almaeen AH, Abdelhafiz AS, Hassan AOS, El Sissy MH. Gabr H, et al. Stem Cell Res Ther. 2020 Sep 10;11(1):390. doi: 10.1186/s13287-020-01876-4. Stem Cell Res Ther. 2020. PMID: 32912325 Free PMC article. - Fanconi anemia gene editing by the CRISPR/Cas9 system.
Osborn MJ, Gabriel R, Webber BR, DeFeo AP, McElroy AN, Jarjour J, Starker CG, Wagner JE, Joung JK, Voytas DF, von Kalle C, Schmidt M, Blazar BR, Tolar J. Osborn MJ, et al. Hum Gene Ther. 2015 Feb;26(2):114-26. doi: 10.1089/hum.2014.111. Hum Gene Ther. 2015. PMID: 25545896 Free PMC article. - Genome editing.
McGrail M, Sakuma T, Bleris L. McGrail M, et al. Sci Rep. 2022 Nov 28;12(1):20497. doi: 10.1038/s41598-022-24850-x. Sci Rep. 2022. PMID: 36443399 Free PMC article. - Bacterial CRISPR/Cas DNA endonucleases: A revolutionary technology that could dramatically impact viral research and treatment.
Kennedy EM, Cullen BR. Kennedy EM, et al. Virology. 2015 May;479-480:213-20. doi: 10.1016/j.virol.2015.02.024. Epub 2015 Mar 7. Virology. 2015. PMID: 25759096 Free PMC article. Review.
References
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. - PubMed
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources