Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex - PubMed (original) (raw)
Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex
Silvija Bilokapic et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Nucleocytoplasmic transport is mediated by nuclear pore complexes (NPCs), enormous assemblies composed of multiple copies of ~30 different proteins called nucleoporins. To unravel the basic scaffold underlying the NPC, we have characterized the species-specific scaffold nucleoporin Nup37 and ELY5/ELYS. Both proteins integrate directly via Nup120/160 into the universally conserved heptameric Y-complex, the critical unit for the assembly and functionality of the NPC. We present the crystal structure of Schizosaccharomyces pombe Nup37 in complex with Nup120, a 174-kDa subassembly that forms one of the two short arms of the Y-complex. Nup37 binds near the bend of the L-shaped Nup120 protein, potentially stabilizing the relative orientation of its two domains. By means of reconstitution assays, we pinpoint residues crucial for this interaction. In vivo and in vitro results show that ELY5 binds near an interface of the Nup120-Nup37 complex. Complementary biochemical and cell biological data refine and consolidate the interactions of Nup120 within the current Y-model. Finally, we propose an orientation of the Y-complex relative to the pore membrane, consistent with the lattice model.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
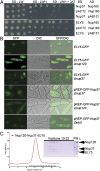
Nup120/160 recruits Nup37 and ELY5/ELYS to the NPC. (A) Y2H interactions between human nucleoporins. Plasmids expressing Lex4 DNA-binding domain (BD) and Gal4 activation domain (AD) fusion constructs of nucleoporins were transformed into a yeast strain, and 10-fold serial dilutions were spotted on synthetic dropout (SD)-Leu-Trp (LW) and SD-Leu-Trp-His (LWH) plates supplemented with 3-amino-1,2,4-triazole (3AT). As a negative control, a combination of the fusion construct and the empty plasmid (pAB151, pACT2) was used. (B) In vivo localization of S. pombe Nup37 and ELY5. Both GFP nucleoporins become distributed throughout the cell in a Δ_nup120_ background (second and fifth rows). GFP-tagged ELY5 and Nup37 are properly targeted to the NPC in Δ_nup37_ and Δ_ely5 S. pombe_ cells (third and sixth rows). (Magnification: 1,000×.) (C) ELY5 forms a complex with Nup120-Nup37 in a gel filtration experiment. Selected fractions (elution volume: 9.85–16.35 mL), marked with a black bar on the chromatogram, were analyzed by SDS/PAGE. The asterisk (*) denotes common impurities. L, loaded proteins; PM, protein marker.
Fig. 2.
Structural analysis of the Nup120–Nup37 complex. (A) Domain structure of Nup120. The propeller domain (PD, blue) with the helical bundle insertion (HB, green) and the irregular α-helical domain (IHD, gray) make the 91 kDa Nup120 NTD. The 39 kDa Nup120 CTD is built from a regular α-helical domain (RHD, red). (B) Cartoon representation of the Nup120-Nup37 heterodimer. The complex comprises full-length nucleoporins with Nup120 colored as in A and Nup37 colored in orange. The β-strands are drawn as arrows, and the α-helices are drawn as cylinders. The two views are rotated by 90° with respect to each other. Nup37 binds close to the bend of the L-shaped Nup120 molecule.
Fig. 3.
Nup120 is structurally distinct from other nucleoporins, composed of a β-propeller domain combined with an α-helical domain. Full-length Nup120 (Left), Nup133 (Center), and Nup851–545 (Right) are gradient-colored blue to white from the N terminus to the C terminus. The structures of the Nup133 β-propeller domain and α-helical domain were determined separately, and are oriented arbitrarily to one another. The ACE1 protein Nup85 in complex with Seh1 (gradient-colored orange to white) exhibits a distinct fold-back structure not observed in Nup120 or Nup133. The Nup85 tail module was modeled and is shown transparently in white.
Fig. 4.
Mutations in Nup37 or Nup120 abolish complex formation. (A) Cartoon representation of Nup37 with elements critically involved in Nup120 binding highlighted in yellow. (B) Gel filtration analysis of Nup37 mutants binding to Nup120 domains shows that the designed mutations abolish the interaction. Complex formation, indicated by an elution peak at ∼11 mL as shown for WT Nup37, is no longer observed for any of the mutants (
Fig. S4_D_
). (C) Ribbon representation of superposed Nup120 NTDs from S. cerevisiae and S. pombe. Differences in the HB region, important for Nup37 binding, are marked. A cartoon representation is used for HB insertion. Nup37 is depicted in the ribbon representation and colored in gray. (D) Cartoon representation of Nup120, with elements critically involved in Nup37 binding highlighted in yellow. (E) Binding data with mutants in Nup120. Gel filtration profiles indicate that the designed mutations in Nup120 abolish interaction with Nup37 (
Fig. S4_E_
).
Fig. 5.
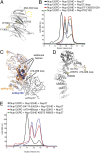
Interaction analysis of the S. pombe Y-complex. (A) S. pombe Nup85-Seh1 stably interacts with Nup145C-Sec13, as judged by the formation of a higher molecular-weight species using size exclusion chromatography (
Fig. S5_C_
). (B) Nup120 binds to preformed Nup85-Seh1-Nup145C-Sec13 tetramer. Addition of Nup37 to this pentameric complex results in the formation of a stable hexameric complex (
Fig. S5_D_
). (C) Nup85 binds Nup120 and Nup145C via its tail module. Binding of the heterodimeric Nup85–Seh1 complex with triple mutation L632D, V632D, and Y636N in the Nup85 tail module (marked as Nup85*-Seh1) to the Y-complex was analyzed by gel filtration. Nup85*-Seh1 binds the Nup145C-Sec13 heterodimer, but incorporation of Nup120 into the tetrameric complex is abolished (
Fig. S7_B_
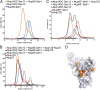
). (D) Threading model of the Nup85 tail module with mapped sequence conservation colored from white to dark orange. Black stars denote the position of the triple mutation used in this study.
Fig. 6.
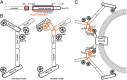
Refined assembly model for the Y-complex. (A) Nup120 regions involved in Nup37 and ELY5 binding are boxed in red and blue, respectively. The domain structure of Nup120 with PD in gray and α-helical regions in black is shown. (B) Relative positions and interactions between the proteins in the Y-complex are shown; the previous consensus model (Left) and the modified model based on the data presented here (Right) are shown. The hub, where the two arms and the stalk of the Y-complex cojoin, has mutual interactions between all extensions rather than Nup120 forming an exclusive tether. The additional proteins Nup37 and ELY5 are recruited via Nup120. Proteins belonging to the ACE1 class are colored in gray. (C) Model of the interaction network within the Nup120–Nup37–ELY5 complex. Black solid arrows depict direct protein–protein interactions, whereas orange arrows indicate proximity to the pore membrane proteins proposed by our work.
Similar articles
- The Role of Nucleoporin Elys in Nuclear Pore Complex Assembly and Regulation of Genome Architecture.
Shevelyov YY. Shevelyov YY. Int J Mol Sci. 2020 Dec 13;21(24):9475. doi: 10.3390/ijms21249475. Int J Mol Sci. 2020. PMID: 33322130 Free PMC article. Review. - Structural evolution of the membrane-coating module of the nuclear pore complex.
Liu X, Mitchell JM, Wozniak RW, Blobel G, Fan J. Liu X, et al. Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):16498-503. doi: 10.1073/pnas.1214557109. Epub 2012 Sep 26. Proc Natl Acad Sci U S A. 2012. PMID: 23019579 Free PMC article. - The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture.
Leksa NC, Brohawn SG, Schwartz TU. Leksa NC, et al. Structure. 2009 Aug 12;17(8):1082-91. doi: 10.1016/j.str.2009.06.003. Epub 2009 Jul 2. Structure. 2009. PMID: 19576787 Free PMC article. - Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly.
Rasala BA, Ramos C, Harel A, Forbes DJ. Rasala BA, et al. Mol Biol Cell. 2008 Sep;19(9):3982-96. doi: 10.1091/mbc.e08-01-0012. Epub 2008 Jul 2. Mol Biol Cell. 2008. PMID: 18596237 Free PMC article. - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review.
Cited by
- Evolutionary divergence of the nuclear pore complex from fungi to metazoans.
Chopra K, Bawaria S, Chauhan R. Chopra K, et al. Protein Sci. 2019 Mar;28(3):571-586. doi: 10.1002/pro.3558. Epub 2018 Dec 24. Protein Sci. 2019. PMID: 30488506 Free PMC article. - Nuclear Import and Export of YAP and TAZ.
Kofler M, Kapus A. Kofler M, et al. Cancers (Basel). 2023 Oct 12;15(20):4956. doi: 10.3390/cancers15204956. Cancers (Basel). 2023. PMID: 37894323 Free PMC article. Review. - A test of double interspecific introgression of nucleoporin genes in Drosophila.
Sawamura K, Maehara K, Keira Y, Ishikawa HO, Sasamura T, Yamakawa T, Matsuno K. Sawamura K, et al. G3 (Bethesda). 2014 Aug 28;4(11):2101-6. doi: 10.1534/g3.114.014027. G3 (Bethesda). 2014. PMID: 25172915 Free PMC article. - In situ structural analysis of the human nuclear pore complex.
von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, Buczak K, Mosalaganti S, Hagen W, Andres-Pons A, Lemke EA, Bork P, Antonin W, Glavy JS, Bui KH, Beck M. von Appen A, et al. Nature. 2015 Oct 1;526(7571):140-143. doi: 10.1038/nature15381. Epub 2015 Sep 23. Nature. 2015. PMID: 26416747 Free PMC article. - The Role of Nucleoporin Elys in Nuclear Pore Complex Assembly and Regulation of Genome Architecture.
Shevelyov YY. Shevelyov YY. Int J Mol Sci. 2020 Dec 13;21(24):9475. doi: 10.3390/ijms21249475. Int J Mol Sci. 2020. PMID: 33322130 Free PMC article. Review.
References
- Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10(3):178–191. - PubMed
- Cook AG, Conti E. Nuclear export complexes in the frame. Curr Opin Struct Biol. 2010;20:247–252. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases