GENCODE: the reference human genome annotation for The ENCODE Project - PubMed (original) (raw)
. 2012 Sep;22(9):1760-74.
doi: 10.1101/gr.135350.111.
Adam Frankish, Jose M Gonzalez, Electra Tapanari, Mark Diekhans, Felix Kokocinski, Bronwen L Aken, Daniel Barrell, Amonida Zadissa, Stephen Searle, If Barnes, Alexandra Bignell, Veronika Boychenko, Toby Hunt, Mike Kay, Gaurab Mukherjee, Jeena Rajan, Gloria Despacio-Reyes, Gary Saunders, Charles Steward, Rachel Harte, Michael Lin, Cédric Howald, Andrea Tanzer, Thomas Derrien, Jacqueline Chrast, Nathalie Walters, Suganthi Balasubramanian, Baikang Pei, Michael Tress, Jose Manuel Rodriguez, Iakes Ezkurdia, Jeltje van Baren, Michael Brent, David Haussler, Manolis Kellis, Alfonso Valencia, Alexandre Reymond, Mark Gerstein, Roderic Guigó, Tim J Hubbard
Affiliations
- PMID: 22955987
- PMCID: PMC3431492
- DOI: 10.1101/gr.135350.111
GENCODE: the reference human genome annotation for The ENCODE Project
Jennifer Harrow et al. Genome Res. 2012 Sep.
Abstract
The GENCODE Consortium aims to identify all gene features in the human genome using a combination of computational analysis, manual annotation, and experimental validation. Since the first public release of this annotation data set, few new protein-coding loci have been added, yet the number of alternative splicing transcripts annotated has steadily increased. The GENCODE 7 release contains 20,687 protein-coding and 9640 long noncoding RNA loci and has 33,977 coding transcripts not represented in UCSC genes and RefSeq. It also has the most comprehensive annotation of long noncoding RNA (lncRNA) loci publicly available with the predominant transcript form consisting of two exons. We have examined the completeness of the transcript annotation and found that 35% of transcriptional start sites are supported by CAGE clusters and 62% of protein-coding genes have annotated polyA sites. Over one-third of GENCODE protein-coding genes are supported by peptide hits derived from mass spectrometry spectra submitted to Peptide Atlas. New models derived from the Illumina Body Map 2.0 RNA-seq data identify 3689 new loci not currently in GENCODE, of which 3127 consist of two exon models indicating that they are possibly unannotated long noncoding loci. GENCODE 7 is publicly available from gencodegenes.org and via the Ensembl and UCSC Genome Browsers.
Figures
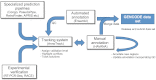
Figure 1.
The GENCODE pipeline. This schematic diagram shows the flow of data between the groups of the GENCODE Consortium. Manual annotation is central to the process but relies on specialized prediction pipelines to provide hints to first-pass annotation and quality control (QC) for completed annotation. Automated annotation supplements manual annotation, the two being merged to produce the GENCODE data set and also to apply QC to the completed annotation. A subset of annotated gene models is subject to experimental validation. The Annotrack tracking system contains data from all groups and is used to highlight differences, coordinate QC, and track outcomes.
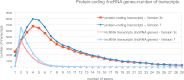
Figure 2.
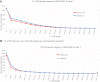
Analysis of exon number of protein-coding and noncoding RNA transcripts. The numbers of exons for each individual transcript annotated at protein-coding and lncRNA loci are plotted for GENCODE 3c (red lines) and GENCODE 7 (blue lines). For each release, darker lines indicate protein-coding transcripts, and lighter lines indicate lncRNA transcripts. The 5′ and 3′ UTR exons of protein-coding transcripts are included.
Figure 3.
A schematic showing the structural annotation of different pseudogene biotypes. The schematic diagram illustrates the categorization of GENCODE pseudogenes on the basis of their origin. Processed pseudogenes are derived by a retrotransposition event and unprocessed pseudogenes by a gene duplication event in both cases, followed by the gain of a disabling mutation. Both processed and unprocessed pseudogenes can retain or gain transcriptional activity, which is reflected in the transcribed_processed and transcribed_unprocessed_pseudogene classification. Polymorphic pseudogenes contain a disabling mutation in the reference genome but are known to be coding in other individuals, while unitary pseudogenes have functional protein-coding orthologs in other species (we have used mouse as a reference) but contain a fixed disabling mutation in human.
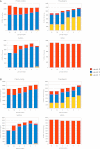
Figure 4.
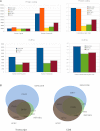
Analysis of GENCODE annotation in 3c through 7. (A) The content of the GENCODE 3c to 7 at the locus level for four broad biotypes: protein-coding, pseudogene, long noncoding RNA (lncRNA), and small RNA (sRNA). The yellow section of each column indicates the proportion of loci classified as Level 1 (validated), the blue part as Level 2 (manually annotated), and the red part as Level 3 (automatically annotated). (B) The analysis of the content of the GENCODE 3c to 7 at the level of the individual transcript. Again, the yellow section of each column indicates the proportion of transcripts classified as Level 1, the blue part as Level 2, and the red part as Level 3.
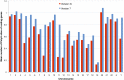
Figure 5.
Comparison of polyA features annotated across all chromosomes. The mean number of polyA features (sites plus signals) for all protein-coding loci are plotted for every chromosome for GENCODE 3c (red columns) and 7 (blue columns).
Figure 6.
Examining the length of 5′ and 3′ UTRs between GENCODE 3c and 7. (A) The length of 5′ UTR sequence (in 50-bp bins) for each protein-coding transcript. 5′ UTR annotation from GENCODE 3c (red) and 7 (blue). (*) A cutoff was made at 949 bases; longer 5′ UTRs do exist. (B) The length of 3′ UTR sequence (in 250-bp bins) for each protein-coding transcript. 3′ UTR annotation from GENCODE 3c (red) and 7 (blue).
Figure 7.
(A) Comparing different publicly available gene sets. The protein-coding content of five major publicly available gene sets— GENCODE, AceView, consensus coding sequence (CCDS), RefSeq, and UCSC—were compared at the level of total gene number, total transcript number, and mean transcripts per locus. (Blue) GENCODE data; (orange) AceView; (yellow) CCDS; (green) RefSeq; (red) UCSC. The lncRNA content of three of these gene sets—GENCODE, RefSeq, and UCSC—were also compared at the level of total gene number, total transcript number, and mean transcripts per locus. Again, GENCODE data are shown in blue, RefSeq in green, and UCSC in red. (B) Overlap between GENCODE, RefSeq, and UCSC at the transcript and CDS levels. Both protein-coding and lncRNA transcripts of all data sets were compared at the transcript level. Two transcripts were considered to match if all their exon junction coordinates were identical in the case of multi-exonic transcripts, or if their transcript coordinates were the same for mono-exonic transcripts. Similarly, the CDSs of two protein-coding transcripts matched when the CDS boundaries and the encompassed exon junctions were identical. Numbers in the intersections involving GENCODE are specific to this data set, otherwise they correspond to any of the other data sets.
Figure 8.
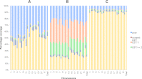
Quality of evidence used to support automatic, manually, and merged annotated transcripts. The level of supporting evidence for automatic only (A), manual only (B), and merged (C) annotated transcripts is shown for each chromosome. (Yellow) The proportion of models with good support; (dark blue) those supported by suspect mRNAs from libraries with known problems with quality; (light green) those with multiple EST support; (orange) those with support from a single EST; (red) those supported by ESTs from suspect libraries; (pale blue) those lacking good support. The number of transcripts across all chromosomes represented in A is 23,855; B, 89,669; and C, 22,535.
Figure 9.
Accessing the GENCODE gene set through UCSC and Ensembl. (A) The composite of screenshots from the UCSC browser shows GENCODE gene annotation displayed in the basic and comprehensive display mode, along with the GENCODE pseudogenes, CCDS models, and a subset of histone modification tracks, DNaseI hypersensitivity clusters, and transcription factor binding site tracks. (B) The configuration display where the user can filter on biotype, annotation method, and transcript type (C). (D) The transcript page in UCSC where the different identifications and version of the transcript can be seen, as well as the evidence used to build the transcript. From the page, the user can click on the Ensembl identification and immediately jump to the Ensembl gene view page (E) and see an overview of the different transcripts in the locus as well as which is a CCDS.
Similar articles
- GENCODE: producing a reference annotation for ENCODE.
Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, Rossier C, Ucla C, Hubbard T, Antonarakis SE, Guigo R. Harrow J, et al. Genome Biol. 2006;7 Suppl 1(Suppl 1):S4.1-9. doi: 10.1186/gb-2006-7-s1-s4. Epub 2006 Aug 7. Genome Biol. 2006. PMID: 16925838 Free PMC article. - GENCODE 2021.
Frankish A, Diekhans M, Jungreis I, Lagarde J, Loveland JE, Mudge JM, Sisu C, Wright JC, Armstrong J, Barnes I, Berry A, Bignell A, Boix C, Carbonell Sala S, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Howe KL, Hunt T, Izuogu OG, Johnson R, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Riera FC, Ruffier M, Schmitt BM, Stapleton E, Suner MM, Sycheva I, Uszczynska-Ratajczak B, Wolf MY, Xu J, Yang YT, Yates A, Zerbino D, Zhang Y, Choudhary JS, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Tress ML, Flicek P. Frankish A, et al. Nucleic Acids Res. 2021 Jan 8;49(D1):D916-D923. doi: 10.1093/nar/gkaa1087. Nucleic Acids Res. 2021. PMID: 33270111 Free PMC article. - GENCODE reference annotation for the human and mouse genomes.
Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, Barnes I, Berry A, Bignell A, Carbonell Sala S, Chrast J, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Hunt T, Izuogu OG, Lagarde J, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Ruffier M, Schmitt BM, Stapleton E, Suner MM, Sycheva I, Uszczynska-Ratajczak B, Xu J, Yates A, Zerbino D, Zhang Y, Aken B, Choudhary JS, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Reymond A, Tress ML, Flicek P. Frankish A, et al. Nucleic Acids Res. 2019 Jan 8;47(D1):D766-D773. doi: 10.1093/nar/gky955. Nucleic Acids Res. 2019. PMID: 30357393 Free PMC article. - EGASP: the human ENCODE Genome Annotation Assessment Project.
Guigó R, Flicek P, Abril JF, Reymond A, Lagarde J, Denoeud F, Antonarakis S, Ashburner M, Bajic VB, Birney E, Castelo R, Eyras E, Ucla C, Gingeras TR, Harrow J, Hubbard T, Lewis SE, Reese MG. Guigó R, et al. Genome Biol. 2006;7 Suppl 1(Suppl 1):S2.1-31. doi: 10.1186/gb-2006-7-s1-s2. Epub 2006 Aug 7. Genome Biol. 2006. PMID: 16925836 Free PMC article. Review. - The Protein-Coding Human Genome: Annotating High-Hanging Fruits.
Hatje K, Mühlhausen S, Simm D, Kollmar M. Hatje K, et al. Bioessays. 2019 Nov;41(11):e1900066. doi: 10.1002/bies.201900066. Epub 2019 Sep 23. Bioessays. 2019. PMID: 31544971 Review.
Cited by
- Spatial genomic heterogeneity within localized, multifocal prostate cancer.
Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY, Zia A, Fox NS, Livingstone J, Shiah YJ, Wang J, Beck TA, Have CL, Chong T, Sam M, Johns J, Timms L, Buchner N, Wong A, Watson JD, Simmons TT, P'ng C, Zafarana G, Nguyen F, Luo X, Chu KC, Prokopec SD, Sykes J, Dal Pra A, Berlin A, Brown A, Chan-Seng-Yue MA, Yousif F, Denroche RE, Chong LC, Chen GM, Jung E, Fung C, Starmans MH, Chen H, Govind SK, Hawley J, D'Costa A, Pintilie M, Waggott D, Hach F, Lambin P, Muthuswamy LB, Cooper C, Eeles R, Neal D, Tetu B, Sahinalp C, Stein LD, Fleshner N, Shah SP, Collins CC, Hudson TJ, McPherson JD, van der Kwast T, Bristow RG. Boutros PC, et al. Nat Genet. 2015 Jul;47(7):736-45. doi: 10.1038/ng.3315. Epub 2015 May 25. Nat Genet. 2015. PMID: 26005866 - LncExpDB: an expression database of human long non-coding RNAs.
Li Z, Liu L, Jiang S, Li Q, Feng C, Du Q, Zou D, Xiao J, Zhang Z, Ma L. Li Z, et al. Nucleic Acids Res. 2021 Jan 8;49(D1):D962-D968. doi: 10.1093/nar/gkaa850. Nucleic Acids Res. 2021. PMID: 33045751 Free PMC article. - Discovery and mass spectrometric analysis of novel splice-junction peptides using RNA-Seq.
Sheynkman GM, Shortreed MR, Frey BL, Smith LM. Sheynkman GM, et al. Mol Cell Proteomics. 2013 Aug;12(8):2341-53. doi: 10.1074/mcp.O113.028142. Epub 2013 Apr 29. Mol Cell Proteomics. 2013. PMID: 23629695 Free PMC article. - Gene Model Annotations for Drosophila melanogaster: Impact of High-Throughput Data.
Matthews BB, Dos Santos G, Crosby MA, Emmert DB, St Pierre SE, Gramates LS, Zhou P, Schroeder AJ, Falls K, Strelets V, Russo SM, Gelbart WM; FlyBase Consortium. Matthews BB, et al. G3 (Bethesda). 2015 Jun 24;5(8):1721-36. doi: 10.1534/g3.115.018929. G3 (Bethesda). 2015. PMID: 26109357 Free PMC article. - Combination of common mtDNA variants results in mitochondrial dysfunction and a connective tissue dysregulation.
Schaefer PM, Scherer Alves L, Lvova M, Huang J, Rathi K, Janssen K, Butic A, Yardeni T, Morrow R, Lott M, Murdock D, Song A, Keller K, Garcia BA, Francomano CA, Wallace DC. Schaefer PM, et al. Proc Natl Acad Sci U S A. 2022 Nov 8;119(45):e2212417119. doi: 10.1073/pnas.2212417119. Epub 2022 Nov 2. Proc Natl Acad Sci U S A. 2022. PMID: 36322731 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- 5U54HG004555/HG/NHGRI NIH HHS/United States
- 095908/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- U54 HG004555/HG/NHGRI NIH HHS/United States
- WT098051/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases