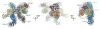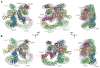Structural basis for a reciprocal regulation between SCF and CSN - PubMed (original) (raw)
Structural basis for a reciprocal regulation between SCF and CSN
Radoslav I Enchev et al. Cell Rep. 2012.
Abstract
Skp1-Cul1-Fbox (SCF) E3 ligases are activated by ligation to the ubiquitin-like protein Nedd8, which is reversed by the deneddylating Cop9 signalosome (CSN). However, CSN also promotes SCF substrate turnover through unknown mechanisms. Through biochemical and electron microscopy analyses, we determined molecular models of CSN complexes with SCF(Skp2/Cks1) and SCF(Fbw7) and found that CSN occludes both SCF functional sites-the catalytic Rbx1-Cul1 C-terminal domain and the substrate receptor. Indeed, CSN binding prevents SCF interactions with E2 enzymes and a ubiquitination substrate, and it inhibits SCF-catalyzed ubiquitin chain formation independent of deneddylation. Importantly, CSN prevents neddylation of the bound cullin, unless binding of a ubiquitination substrate triggers SCF dissociation and neddylation. Taken together, the results provide a model for how reciprocal regulation sensitizes CSN to the SCF assembly state and inhibits a catalytically competent SCF until a ubiquitination substrate drives its own degradation by displacing CSN, thereby promoting cullin neddylation and substrate ubiquitination.
Copyright © 2012 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
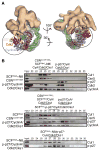
Figure 1
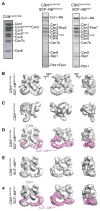
Reconstitution and single particle Electron Microscope analysis of CSN-SCF complexes. (A) Coomassie-stained SDS-PAGE of recombinant CSNCsn5H138A and reconstituted CSN-SCF complexes after gel filtration. (B – F) Surface views of EM density maps of (B) CSNCsn5H138A-SCF~N8Skp2/Cks1, (C) CSNCsn5H138A, (D) CSNCsn5H138A-SCF~N8Skp2/Cks1 segmented into its SCF~N8Skp2/Cks1 (purple) and CSN (grey) subcomplexes. (E) CSNCsn5H138A-SCF~N8Fbw7, (F) CSNCsn5H138A-SCF~N8Fbw7 segmented into its CSNCsn5H138A-SCF~N8Fbw7 (purple) and CSN (grey) subcomplexes. See also Figures S1 and S3.
Figure 2
Molecular model for CSN. The CSN segment from the CSNCsn5H138A-SCF~N8Skp2/Cks1 map is shown as grey mesh. Left - PCI cluster side view. A dotted arc and color-coded arrows indicate the approximately coplanar positions of the winged-helix domains. MPN subunits are omitted for clarity. Center - opposite side, characterized by a protrusion formed by the two MPN domain subunits, Csn 5 and 6. Right - edge-on to the coplanar PCI cluster. The protrusion formed by the Ccn5 and 6 MPN subunits is left of the PCI cluster. Csn6 is more closely integrated with the PCI cluster while Csn5 is angled away. See also Figure S2.
Figure 3
Molecular models for (A) CSNCsn5H138A-SCF~N8Skp2/Cks1 and (B) CSNCsn5H138A-SCF~N8Fbw7. Brown and purple (A) or purple (B) arrows in the central views indicate contact points between the SCF substrate receptors and CSN. Black arrows in the right hand views indicate the basic canyon region of Cul1. See also Figures S1 and S3.
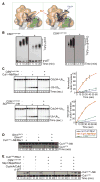
Figure 4
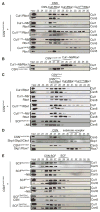
CSN-SCF interactions analyzed by analytical size exclusion chromatography. Indicated SCF complexes were tested with equimolar amounts of (A, D, E) CSN harboring the Csn5H138A active site mutation, (B) CSN harboring an N-terminally truncated Csn2 and Csn5H138A or (C) a CSN complex lacking Csn5. Input and peak fractions (numbered) were blotted with antibodies indicated to the right. Fractions in which particular complexes were eluted are indicated above each panel. Neddylated complexes are labeled with ~N8. See also Figure S4.
Figure 5
CSN-SCF interactions in the presence of ubiquitination substrate. (A) Views of the CSNCsn5H138A-SCF~N8Skp2/Cks1 map are shown with CSN as an orange surface, SCFSkp2/Cks1~N8 as a gray mesh and the atomic coordinates as in Figure 3A. Docked Cdk2 (red), CyclinA (blue) and N- and C-terminal segments of p-p27 (yellow) are indicated by circles. A potential steric clash of p-p27 with the CSN density is indicated by a dashed yellow curve and an arrow on the left. (B) Neddylated (~N8) or non-neddylated SCFSkp2 complexes were incubated with CyclinA/Cdk2/Cks1 and p-p27, in the presence or absence of equimolar amounts of deneddylation-defective CSNCsn5H138A and analyzed by size exclusion chromatography. See also Figure S5.
Figure 6
CSN-SCF binding regulates ubiquitination and neddylation. (A) Zoomed-in surface view of CSNCsn5H138A-SCF~N8Skp2/Cks1, color-coded as Figure 5A, showing overlay of all reported Rbx1 conformations without (left) or with docked Cdc34 model (right). Cul1411–690 (CTD without Helix29 and WHB) shown in green, Rbx1 and Cds34 orientations shown as hues of red and blue. (B) In vitro ubiquitination of p-p27 (left) CyclinEphospho-peptide (right),assayed in the absence (−) or presence (+) of catalytically-inactive CSNCsn5H138A. Unmodified and poly-ubiquitinated substrates (−(Ub)n) detected by immunoblotting with p-p27- and biotin-antibodies respectively. (C) Pulse-chase [32P]~ubiquitin (Ub) transfer from Cdc34 to lysine-less ubiquitin (UbK0) in the presence (+) or absence (−) of CSNCsn5H138A and neddylated Cul1/Rbx1 (upper panel) or neddylated SCFSkp2/Cks1 (lower panel). Formation of di-Ub was assayed by autoradiography (left hand panels), and quantified by plotting the percentage (%) of di-Ub formation as a function of time (right hand panels). (D) Deneddylation of Cul1CTD~N8/Rbx1 by CSN assayed as described in Figure S6B in the presence (+) or absence (−) of wild type (WT) or Rbx1-interction deffective R547A mutant Glmn. (E) Neddylation (~N8) of Cul1SCE/Rbx1 measured by immunoblotting with Cul1 antibodies, in the presence (+) or absence (−) of CSNCsn5H138A, Skp1/Skp2/Cks1 and CyclinA/Cdk2 and p-p27. See also Figures S6 and S7.
Similar articles
- HSC70 coordinates COP9 signalosome and SCF ubiquitin ligase activity to enable a prompt stress response.
Nishimura S, Kioka H, Ding S, Hakui H, Shinomiya H, Tanabe K, Hitsumoto T, Matsuoka K, Kato H, Tsukamoto O, Asano Y, Takashima S, Enchev RI, Sakata Y. Nishimura S, et al. EMBO Rep. 2025 Feb 6. doi: 10.1038/s44319-025-00376-x. Online ahead of print. EMBO Rep. 2025. PMID: 39915298 - Substrate phosphorylation and feedback regulation in JFK-promoted p53 destabilization.
Sun L, Shi L, Wang F, Huangyang P, Si W, Yang J, Yao Z, Shang Y. Sun L, et al. J Biol Chem. 2011 Feb 11;286(6):4226-35. doi: 10.1074/jbc.M110.195115. Epub 2010 Dec 2. J Biol Chem. 2011. PMID: 21127074 Free PMC article. - Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Horn-Ghetko D, Krist DT, Prabu JR, Baek K, Mulder MPC, Klügel M, Scott DC, Ovaa H, Kleiger G, Schulman BA. Horn-Ghetko D, et al. Nature. 2021 Feb;590(7847):671-676. doi: 10.1038/s41586-021-03197-9. Epub 2021 Feb 3. Nature. 2021. PMID: 33536622 Free PMC article. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Control of cell growth by the SCF and APC/C ubiquitin ligases.
Skaar JR, Pagano M. Skaar JR, et al. Curr Opin Cell Biol. 2009 Dec;21(6):816-24. doi: 10.1016/j.ceb.2009.08.004. Epub 2009 Sep 21. Curr Opin Cell Biol. 2009. PMID: 19775879 Free PMC article. Review.
Cited by
- Hepatic deficiency of COP9 signalosome subunit 8 induces ubiquitin-proteasome system impairment and Bim-mediated apoptosis in murine livers.
Lei D, Li F, Su H, Liu J, Wei N, Wang X. Lei D, et al. PLoS One. 2013 Jul 1;8(7):e67793. doi: 10.1371/journal.pone.0067793. Print 2013. PLoS One. 2013. PMID: 23840878 Free PMC article. - Activity-based profiling of cullin-RING E3 networks by conformation-specific probes.
Henneberg LT, Singh J, Duda DM, Baek K, Yanishevski D, Murray PJ, Mann M, Sidhu SS, Schulman BA. Henneberg LT, et al. Nat Chem Biol. 2023 Dec;19(12):1513-1523. doi: 10.1038/s41589-023-01392-5. Epub 2023 Aug 31. Nat Chem Biol. 2023. PMID: 37653169 Free PMC article. - Basis for metabolite-dependent Cullin-RING ligase deneddylation by the COP9 signalosome.
Lin H, Zhang X, Liu L, Fu Q, Zang C, Ding Y, Su Y, Xu Z, He S, Yang X, Wei X, Mao H, Cui Y, Wei Y, Zhou C, Du L, Huang N, Zheng N, Wang T, Rao F. Lin H, et al. Proc Natl Acad Sci U S A. 2020 Feb 25;117(8):4117-4124. doi: 10.1073/pnas.1911998117. Epub 2020 Feb 11. Proc Natl Acad Sci U S A. 2020. PMID: 32047038 Free PMC article. - Dynamic regulation of the COP9 signalosome in response to DNA damage.
Füzesi-Levi MG, Ben-Nissan G, Bianchi E, Zhou H, Deery MJ, Lilley KS, Levin Y, Sharon M. Füzesi-Levi MG, et al. Mol Cell Biol. 2014 Mar;34(6):1066-76. doi: 10.1128/MCB.01598-13. Epub 2014 Jan 13. Mol Cell Biol. 2014. PMID: 24421388 Free PMC article. - SCF(FBXW7α) modulates the intra-S-phase DNA-damage checkpoint by regulating Polo like kinase-1 stability.
Giráldez S, Herrero-Ruiz J, Mora-Santos M, Japón MÁ, Tortolero M, Romero F. Giráldez S, et al. Oncotarget. 2014 Jun 30;5(12):4370-83. doi: 10.18632/oncotarget.2021. Oncotarget. 2014. PMID: 24970797 Free PMC article.
References
- Chew EH, Hagen T. Substrate-mediated regulation of cullin neddylation. J Biol Chem. 2007;282:17032–17040. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHMI_/Howard Hughes Medical Institute/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- R01 GM069530/GM/NIGMS NIH HHS/United States
- R01GM069530/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous