Ancient admixture in human history - PubMed (original) (raw)
Ancient admixture in human history
Nick Patterson et al. Genetics. 2012 Nov.
Abstract
Population mixture is an important process in biology. We present a suite of methods for learning about population mixtures, implemented in a software package called ADMIXTOOLS, that support formal tests for whether mixture occurred and make it possible to infer proportions and dates of mixture. We also describe the development of a new single nucleotide polymorphism (SNP) array consisting of 629,433 sites with clearly documented ascertainment that was specifically designed for population genetic analyses and that we genotyped in 934 individuals from 53 diverse populations. To illustrate the methods, we give a number of examples that provide new insights about the history of human admixture. The most striking finding is a clear signal of admixture into northern Europe, with one ancestral population related to present-day Basques and Sardinians and the other related to present-day populations of northeast Asia and the Americas. This likely reflects a history of admixture between Neolithic migrants and the indigenous Mesolithic population of Europe, consistent with recent analyses of ancient bones from Sweden and the sequencing of the genome of the Tyrolean "Iceman."
Figures
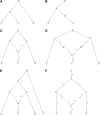
Figure 1
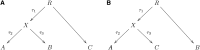
_f_-statistics: (A) A simple phylogenetic tree, (B) the additivity of branch lengths; the genetic drift between (A, B) computed using our _f_-statistic-based methods is the same as the sum of the genetic drifts between (A, B) and (B, C), regardless of the population in which SNPs are ascertained, (C) phylogenetic tree with simple admixture, (D) a more general form of Figure 1C, (E) example of an outgroup case, and (F) example of admixture with an outgroup.
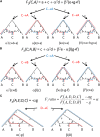
Figure 2
Visual computation of expected values of F2, F3, and F4 statistics. See Appendix 2 for a discussion of this figure.
Figure 3
_D_-statistics provide formal tests for whether an unrooted phylogenetic tree applies to the data, assuming that the analyzed SNPs are ascertained as polymorphic in a population that is an outgroup to both populations (Y, Z) that make up one of the clades. (A) A simple unrooted phylogeny, (B) phylogenies in which (Y, Z) and (W, X) are clades that diverge from a common root, (C) phylogenies in which (Y, Z) are a clade and W and X are increasingly distant outgroups, and (D) a phylogeny to test if human Eurasian populations (A, B) form a clade with respect to sub-Saharan Africans (Yoruba).
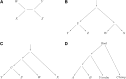
Figure 4
A phylogeny explaining _f_4-ratio estimation.
Figure 5
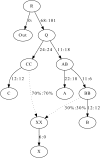
Admixture graph fitting: We show an admixture graph fitted by qpGraph for simulated data. We simulated 50,000 unlinked SNPs ascertained as heterozygous in a single diploid individual from the outgroup Out. Sample sizes were 50 in all populations and the historical population sizes were all taken to be 10,000. The true values of parameters are before the colon and the estimated values afterward. Mixture proportions are given as percentages, and branch lengths are given in units of _F_st (before the colon) and _f_2 values (after). _F_2 and _F_st are multiplied by 1000. The fitted admixture weights are exact, up to the resolution shown, while the match of branch lengths to the truth is rather approximate.
Figure 6
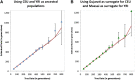
rolloff simulation results: We simulated data for 100 individuals of 20% European and 80% African ancestry, where the mixture occurred between 50 and 800 generations ago. Phased data from HapMap3 CEU and YRI populations was used for the simulations. We performed rolloff analysis using CEU and YRI (A) and using Gujarati and Maasai (B) as reference populations. We plot the true date of mixture (dotted line) against the estimated date computed by rolloff (points in blue A and green B). Standard errors were calculated using the weighted block jackknife. To test the bias in the estimated dates, we repeated each simulation 10 times. The estimated date based on the 10 simulations is shown in red.
Figure 7
rolloff analysis of real data: We applied rolloff to compute admixture LD between all pairs of markers in each admixed population. We plot the correlation as a function of genetic distance for (A) Xhosa, (B) Uygur, (C) Spain, (D) Greece, and (E) CEU and French. The title of each includes information about the reference populations that were used for the analysis. We fit an exponential distribution to the output of rolloff to estimate the date of the mixture (estimated dates ±SE shown in years). We do not show inter-SNP intervals of <0.5 cM as we have found that at this distance admixture LD begins to be confounded by background LD.
Figure 8
Bell-Beaker culture. On the left we show some Beaker culture objects (from Bruchsal City Museum). On the right we show a map of Bell-Beaker attested sites. We are grateful to Thomas Ihle for the Bruchsal Museum photograph. It is licensed under the Creative Commons Attribution-Share Alike 3.0 Unported license, and a GNU Free documentation license. The map is public domain, licensed under a creative commons license, and adapted from a map in Harrison (1980).
Figure 9
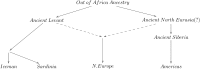
Northeast Asian-related admixture in northern Europe. A proposed model of population relationships that can explain some features observed in our genetic data.
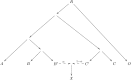
Figure C1
(A) Appendix C, Theorem 1. (B) Appendix C, Theorem 2.
Similar articles
- Efficient moment-based inference of admixture parameters and sources of gene flow.
Lipson M, Loh PR, Levin A, Reich D, Patterson N, Berger B. Lipson M, et al. Mol Biol Evol. 2013 Aug;30(8):1788-802. doi: 10.1093/molbev/mst099. Epub 2013 May 24. Mol Biol Evol. 2013. PMID: 23709261 Free PMC article. - Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe.
Sikora M, Carpenter ML, Moreno-Estrada A, Henn BM, Underhill PA, Sánchez-Quinto F, Zara I, Pitzalis M, Sidore C, Busonero F, Maschio A, Angius A, Jones C, Mendoza-Revilla J, Nekhrizov G, Dimitrova D, Theodossiev N, Harkins TT, Keller A, Maixner F, Zink A, Abecasis G, Sanna S, Cucca F, Bustamante CD. Sikora M, et al. PLoS Genet. 2014 May 8;10(5):e1004353. doi: 10.1371/journal.pgen.1004353. eCollection 2014 May. PLoS Genet. 2014. PMID: 24809476 Free PMC article. - Genetic relatedness of indigenous ethnic groups in northern Borneo to neighboring populations from Southeast Asia, as inferred from genome-wide SNP data.
Yew CW, Hoque MZ, Pugh-Kitingan J, Minsong A, Voo CLY, Ransangan J, Lau STY, Wang X, Saw WY, Ong RT, Teo YY, Xu S, Hoh BP, Phipps ME, Kumar SV. Yew CW, et al. Ann Hum Genet. 2018 Jul;82(4):216-226. doi: 10.1111/ahg.12246. Epub 2018 Mar 9. Ann Hum Genet. 2018. PMID: 29521412 - Review of the Forensic Applicability of Biostatistical Methods for Inferring Ancestry from Autosomal Genetic Markers.
Tvedebrink T. Tvedebrink T. Genes (Basel). 2022 Jan 14;13(1):141. doi: 10.3390/genes13010141. Genes (Basel). 2022. PMID: 35052480 Free PMC article. Review. - African genetic diversity provides novel insights into evolutionary history and local adaptations.
Choudhury A, Aron S, Sengupta D, Hazelhurst S, Ramsay M. Choudhury A, et al. Hum Mol Genet. 2018 Aug 1;27(R2):R209-R218. doi: 10.1093/hmg/ddy161. Hum Mol Genet. 2018. PMID: 29741686 Free PMC article. Review.
Cited by
- High levels of consanguinity in a child from Paquimé, Chihuahua, Mexico.
Sedig J, Snow M, Searcy M, Diaz JLP, LeBlanc S, Ramos F, Eccles L, Reich D. Sedig J, et al. Antiquity. 2024 Aug;98(400):1023-1039. doi: 10.15184/aqy.2024.94. Epub 2024 Aug 13. Antiquity. 2024. PMID: 39493412 Free PMC article. - The rise and transformation of Bronze Age pastoralists in the Caucasus.
Ghalichi A, Reinhold S, Rohrlach AB, Kalmykov AA, Childebayeva A, Yu H, Aron F, Semerau L, Bastert-Lamprichs K, Belinskiy AB, Berezina NY, Berezin YB, Broomandkhoshbacht N, Buzhilova AP, Erlikh VR, Fehren-Schmitz L, Gambashidze I, Kantorovich AR, Kolesnichenko KB, Lordkipanidze D, Magomedov RG, Malek-Custodis K, Mariaschk D, Maslov VE, Mkrtchyan L, Nagler A, Fazeli Nashli H, Ochir M, Piotrovskiy YY, Saribekyan M, Sheremetev AG, Stöllner T, Thomalsky J, Vardanyan B, Posth C, Krause J, Warinner C, Hansen S, Haak W. Ghalichi A, et al. Nature. 2024 Oct 30. doi: 10.1038/s41586-024-08113-5. Online ahead of print. Nature. 2024. PMID: 39478221 - Biomolecular analysis of the Epigravettian human remains from Riparo Tagliente in northern Italy.
Yavuz OE, Oxilia G, Silvestrini S, Tassoni L, Reiter E, Drucker DG, Talamo S, Fontana F, Benazzi S, Posth C. Yavuz OE, et al. Commun Biol. 2024 Oct 30;7(1):1415. doi: 10.1038/s42003-024-06979-9. Commun Biol. 2024. PMID: 39478147 Free PMC article. - Archaeogenetic analysis revealed East Eurasian paternal origin to the Aba royal family of Hungary.
Varga GIB, Maróti Z, Schütz O, Maár K, Nyerki E, Tihanyi B, Váradi OA, Gînguță A, Kovács B, Kiss P, Dosztig M, Gallina Z, Török T, Szabó JB, Makoldi M, Neparáczki E. Varga GIB, et al. iScience. 2024 Sep 14;27(10):110892. doi: 10.1016/j.isci.2024.110892. eCollection 2024 Oct 18. iScience. 2024. PMID: 39474080 Free PMC article.
References
- Anthony D. W., 2007. The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World, Princeton University Press, Princeton, NJ.
- Barnard A., 1992. Hunters and Herders of Southern Africa. A comparative ethnography of the Khoisan peoples, Cambridge University Press, Cambridge, UK.
- Bramanti B., Thomas M. G., Haak W., Unterlaender M., Jores P., et al. 2009. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science 326: 137–140. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources