Stitching gene fragments with a network matching algorithm improves gene assembly for metagenomics - PubMed (original) (raw)
Stitching gene fragments with a network matching algorithm improves gene assembly for metagenomics
Yu-Wei Wu et al. Bioinformatics. 2012.
Abstract
Motivation: One of the difficulties in metagenomic assembly is that homologous genes from evolutionarily closely related species may behave like repeats and confuse assemblers. As a result, small contigs, each representing a short gene fragment, instead of complete genes, may be reported by an assembler. This further complicates annotation of metagenomic datasets, as annotation tools (such as gene predictors or similarity search tools) typically perform poorly on configs encoding short gene fragments.
Results: We present a novel way of using the de Bruijn graph assembly of metagenomes to improve the assembly of genes. A network matching algorithm is proposed for matching the de Bruijn graph of contigs against reference genes, to derive 'gene paths' in the graph (sequences of contigs containing gene fragments) that have the highest similarities to known genes, allowing gene fragments contained in multiple contigs to be connected to form more complete (or intact) genes. Tests on simulated and real datasets show that our approach (called GeneStitch) is able to significantly improve the assembly of genes from metagenomic sequences, by connecting contigs with the guidance of homologous genes-information that is orthogonal to the sequencing reads. We note that the improvement of gene assembly can be observed even when only distantly related genes are available as the reference. We further propose to use 'gene graphs' to represent the assembly of reads from homologous genes and discuss potential applications of gene graphs to improving functional annotation for metagenomics.
Availability: The tools are available as open source for download at http://omics.informatics.indiana.edu/GeneStitch
Contact: yye@indiana.edu.
Figures
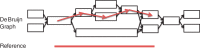
Fig. 1.
Alignment between a de Bruijn graph and a reference sequence. Blocks in the de Bruijn graph represent nodes, and black arrowheads represent the directed edges that connect nodes with overlapping k − 1 mers. Typically, a de Bruijn graph-based assembler will output each of the nodes as a contig. Red arrowheads constitute the optimal path of the nodes that aligns with the reference sequence derived by the network matching algorithm
Fig. 2.
Improvement of gene assembly by GeneStitch for the simulated and real community datasets, as evaluated by gene coverage (A) and the number of complete genes (B)
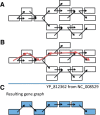
Fig. 3.
An example demonstrating the inference of a gene path from a connected component in the de Bruijn graph. The reference gene recruited by BLAST in this example is YP_812362. (A) In total, 17 nodes are present in this connected component. (B) The path found by GeneStitch using the reference gene. (C) The gene path
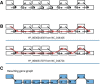
Fig. 4.
An example demonstrating the construction of a gene graph by merging gene paths. (A) only 19 nodes are shown in this figure for clarity (the actual component is larger). (B) Two paths are found by GeneStitch, using YP_003601430 and YP_004031707 as the reference genes. (C) The two paths are merged into a gene graph
Similar articles
- Utilizing de Bruijn graph of metagenome assembly for metatranscriptome analysis.
Ye Y, Tang H. Ye Y, et al. Bioinformatics. 2016 Apr 1;32(7):1001-8. doi: 10.1093/bioinformatics/btv510. Epub 2015 Aug 29. Bioinformatics. 2016. PMID: 26319390 Free PMC article. - MegaGTA: a sensitive and accurate metagenomic gene-targeted assembler using iterative de Bruijn graphs.
Li D, Huang Y, Leung CM, Luo R, Ting HF, Lam TW. Li D, et al. BMC Bioinformatics. 2017 Oct 16;18(Suppl 12):408. doi: 10.1186/s12859-017-1825-3. BMC Bioinformatics. 2017. PMID: 29072142 Free PMC article. - GraphBin: refined binning of metagenomic contigs using assembly graphs.
Mallawaarachchi V, Wickramarachchi A, Lin Y. Mallawaarachchi V, et al. Bioinformatics. 2020 Jun 1;36(11):3307-3313. doi: 10.1093/bioinformatics/btaa180. Bioinformatics. 2020. PMID: 32167528 - Assessment of metagenomic assemblers based on hybrid reads of real and simulated metagenomic sequences.
Wang Z, Wang Y, Fuhrman JA, Sun F, Zhu S. Wang Z, et al. Brief Bioinform. 2020 May 21;21(3):777-790. doi: 10.1093/bib/bbz025. Brief Bioinform. 2020. PMID: 30860572 Free PMC article. Review. - MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices.
Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, Yamashita H, Lam TW. Li D, et al. Methods. 2016 Jun 1;102:3-11. doi: 10.1016/j.ymeth.2016.02.020. Epub 2016 Mar 21. Methods. 2016. PMID: 27012178 Review.
Cited by
- Identifying similar transcripts in a related organism from de Bruijn graphs of RNA-Seq data, with applications to the study of salt and waterlogging tolerance in Melilotus.
Fu S, Chang PL, Friesen ML, Teakle NL, Tarone AM, Sze SH. Fu S, et al. BMC Genomics. 2019 Jun 6;20(Suppl 5):425. doi: 10.1186/s12864-019-5702-5. BMC Genomics. 2019. PMID: 31167652 Free PMC article. - A scalable and accurate targeted gene assembly tool (SAT-Assembler) for next-generation sequencing data.
Zhang Y, Sun Y, Cole JR. Zhang Y, et al. PLoS Comput Biol. 2014 Aug 14;10(8):e1003737. doi: 10.1371/journal.pcbi.1003737. eCollection 2014 Aug. PLoS Comput Biol. 2014. PMID: 25122209 Free PMC article. - ORFograph: search for novel insecticidal protein genes in genomic and metagenomic assembly graphs.
Dvorkina T, Bankevich A, Sorokin A, Yang F, Adu-Oppong B, Williams R, Turner K, Pevzner PA. Dvorkina T, et al. Microbiome. 2021 Jun 28;9(1):149. doi: 10.1186/s40168-021-01092-z. Microbiome. 2021. PMID: 34183047 Free PMC article. - Music of metagenomics-a review of its applications, analysis pipeline, and associated tools.
Wajid B, Anwar F, Wajid I, Nisar H, Meraj S, Zafar A, Al-Shawaqfeh MK, Ekti AR, Khatoon A, Suchodolski JS. Wajid B, et al. Funct Integr Genomics. 2022 Feb;22(1):3-26. doi: 10.1007/s10142-021-00810-y. Epub 2021 Oct 18. Funct Integr Genomics. 2022. PMID: 34657989 Review. - Identification and Resolution of Microdiversity through Metagenomic Sequencing of Parallel Consortia.
Nelson WC, Maezato Y, Wu YW, Romine MF, Lindemann SR. Nelson WC, et al. Appl Environ Microbiol. 2015 Oct 23;82(1):255-67. doi: 10.1128/AEM.02274-15. Print 2016 Jan 1. Appl Environ Microbiol. 2015. PMID: 26497460 Free PMC article.
References
- Bentley D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials