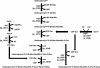Lactobacillus plantarum possesses the capability for wall teichoic acid backbone alditol switching - PubMed (original) (raw)
Lactobacillus plantarum possesses the capability for wall teichoic acid backbone alditol switching
Peter A Bron et al. Microb Cell Fact. 2012.
Abstract
Background: Specific strains of Lactobacillus plantarum are marketed as health-promoting probiotics. The role and interplay of cell-wall compounds like wall- and lipo-teichoic acids (WTA and LTA) in bacterial physiology and probiotic-host interactions remain obscure. L. plantarum WCFS1 harbors the genetic potential to switch WTA backbone alditol, providing an opportunity to study the impact of WTA backbone modifications in an isogenic background.
Results: Through genome mining and mutagenesis we constructed derivatives that synthesize alternative WTA variants. The mutants were shown to completely lack WTA, or produce WTA and LTA that lack D-Ala substitution, or ribitol-backbone WTA instead of the wild-type glycerol-containing backbone. DNA micro-array experiments established that the tarIJKL gene cluster is required for the biosynthesis of this alternative WTA backbone, and suggest ribose and arabinose are precursors thereof. Increased tarIJKL expression was not observed in any of our previously performed DNA microarray experiments, nor in qRT-PCR analyses of L. plantarum grown on various carbon sources, leaving the natural conditions leading to WTA backbone alditol switching, if any, to be identified. Human embryonic kidney NF-κB reporter cells expressing Toll like receptor (TLR)-2/6 were exposed to purified WTAs and/or the TA mutants, indicating that WTA is not directly involved in TLR-2/6 signaling, but attenuates this signaling in a backbone independent manner, likely by affecting the release and exposure of immunomodulatory compounds such as LTA. Moreover, human dendritic cells did not secrete any cytokines when purified WTAs were applied, whereas they secreted drastically decreased levels of the pro-inflammatory cytokines IL-12p70 and TNF-α after stimulation with the WTA mutants as compared to the wild-type.
Conclusions: The study presented here correlates structural differences in WTA to their functional characteristics, thereby providing important information aiding to improve our understanding of molecular host-microbe interactions and probiotic functionality.
Figures
Figure 1
Functions of the proteins encoded by the tag and tar genes in the biosynthesis of poly(Gro-P)- and poly(Rbo-P)-containing WTA backbones, respectively [adapted from [9]. Numbers between brackets indicate the gene-identifiers of tag and tar homologues in the L. plantarum WCFS1 genome and Rib represents ribulose. Regardless of the backbone type, the biosynthesis of WTA is initiated with the consecutive coupling of UDP-activated _N_-acetylglucosamine (Glc_N_ac) and _N_-acetylmannosamine (Man_N_ac) to undecaprenyl phosphate on the cytoplasmic side of the cell membrane by TagO and TagA, respectively [5]. Gro-P is CDP-activated by TagD and coupled to this disaccharide linkage unit by the primase TagB. In strains producing poly(Gro-P) WTA the final biosynthetic step involves the coupling of a large but variable amount of CDP-activated Gro-P monomers by the oligomerase TagF. Although TagF homologues (sometimes designated TarF) are present in several strains producing poly(Rbo-P) WTA, these enzymes are generally smaller than their TagF counterparts and their activity is limited to the incorporation of 1 or 2 additional CDP-activated Gro-P monomers [9,62]. In the final biosynthetic steps CDP-activated Rbo-P, produced by TarJ and TarI, is added to the Gro-P mono-, di- or tri-mer by the primase (TarK) and the oligomerase activity (TarL), resulting in the addition of multiple repeating units of Rbo-P [5,9,13,63]. Following its synthesis, the complete WTA polymer is proposed to be transported across the cytoplasmic membrane by the _tagGH_-encoded ABC transporter, and is subsequently coupled to the 6-hydroxyl of Mur_N_Ac in peptidoglycan by an unknown transferase [5,13]. These basic WTA polymers can be substituted with D-alanyl esters through the activity of the enzymes encoded in the dlt operon [31,64,65] and/or glycosyl moieties by homologues of TagE or TarM of B. subtilis and S. aureus, respectively [39,40].
Figure 2
Chromosomal organization of the 7 distinct genetic loci (represented by the iterating grey- and unshaded boxes) harboring the tag and tar genes in the L. plantarum WCFS1 and JDM-1 genomes (panel A). The tag and tar genes are highlighted in bold and named according to the current annotation in the WCFS1 genome. Colors represent different functional classes as defined by the microbial genome viewer tool [66]. Panel B displays detailed sequence information around the tagD1-tagF1-tagF2 locus (last grey-shaded box in Panel A) with the start codon of the downstream-encoded lp_0271 (vdcB) underscored.
Figure 3
Sugars, alditols, and D-Ala composition of WTA isolated from L. plantarum WCFS1 (I), and the dltX-D (II), and tagF1-F2 (III) mutants. Panel A represents chromatograms on a Shodex NH2P-50 4E column, whereas panel B displays chromatograms on a Shodex SUGAR SC1011 column. Using pure compounds, peaks 1, 2, 3, 4 and 5 were identified to represent anhydroribitol, glycerol, ribitol, glucose, and alanine, respectively. The experiment is representative of 2 HPLC analyses.
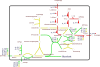
Figure 4
Simpheny visualization map (www. genomatica.com) using the L. plantarum specific model [67]of the biosynthetic pathways involved in WTA backbone precursor biosynthesis. Red and green arrow(head) represents induced and repressed expression levels as assessed by DNA microarrays, whereas yellow arrows and boxes indicate unchanged expression levels in the L. plantarum tagF1-2 mutant as compared to the wild-type, respectively. The size of the arrowheads reflects the quantitative induction/repression levels, the black line represents the cytoplasmic membrane, and [e] indicates extracellular localization.
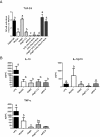
Figure 5
Immunomodulating and signaling capacity of the L. plantarum TA mutants. Panel A displays NF-κB pathway activation relative to L. plantarum WCFS1, as measured by a luminescence reporter in HEK cell lines expressing TLR-2/6 after exposure to PAM3CSK (positive control), purified WTA (5.0 μg/ml, see Additional file 4: Figure S2 and Additional file 5: Figure S3 for concentration ranges) and/or L. plantarum WCFS1, the tagO, _tagF1_-2, or dltX-D mutants. The ratios of HEK reporter to bacterial cell were 1:15. All strains were grown in triplicate and each culture was applied three times to the assay. Bars represent averages of these nine measurements with standard deviations. Letters a-d represent classes of statistically different responses as determined by ANOVA multiple comparison. Panel B displays cytokine levels secreted by monocyte derived iDCs stimulated with LPS (positive control), L. plantarum WCFS1, and the tagO, _tagF1_-2 and dltX-D mutants. The ratios of dendritic to bacterial cells were 1:10. All strains were grown in duplicate and each culture was applied in duplicate to the assay. Bars represent averages of these four measurements with standard deviations. Individual letters (a-c) represent classes of statistically significant different responses as determined by restricted maximum likelihood analysis, whereas “bc” represents a class not significantly different from “b” nor “c”.
Similar articles
- Characterization of the transcriptional regulation of the tarIJKL locus involved in ribitol-containing wall teichoic acid biosynthesis in Lactobacillus plantarum.
Tomita S, Lee IC, van Swam II, Boeren S, Vervoort J, Bron PA, Kleerebezem M. Tomita S, et al. Microbiology (Reading). 2016 Feb;162(2):420-432. doi: 10.1099/mic.0.000229. Epub 2015 Dec 17. Microbiology (Reading). 2016. PMID: 26678992 - Determination of strain-specific wall teichoic acid structures in Lactobacillus plantarum reveals diverse α-D-glucosyl substitutions and high structural uniformity of the repeating units.
Tomita S, Furihata K, Tanaka N, Satoh E, Nukada T, Okada S. Tomita S, et al. Microbiology (Reading). 2012 Nov;158(Pt 11):2712-2723. doi: 10.1099/mic.0.060913-0. Epub 2012 Aug 23. Microbiology (Reading). 2012. PMID: 22918894 - The impact of Lactobacillus plantarum WCFS1 teichoic acid D-alanylation on the generation of effector and regulatory T-cells in healthy mice.
Smelt MJ, de Haan BJ, Bron PA, van Swam I, Meijerink M, Wells JM, Kleerebezem M, Faas MM, de Vos P. Smelt MJ, et al. PLoS One. 2013 Apr 30;8(4):e63099. doi: 10.1371/journal.pone.0063099. Print 2013. PLoS One. 2013. PMID: 23646181 Free PMC article. - Controlling biofilm and virulence properties of Gram-positive bacteria by targeting wall teichoic acid and lipoteichoic acid.
Jeong GJ, Khan F, Tabassum N, Cho KJ, Kim YM. Jeong GJ, et al. Int J Antimicrob Agents. 2023 Oct;62(4):106941. doi: 10.1016/j.ijantimicag.2023.106941. Epub 2023 Aug 2. Int J Antimicrob Agents. 2023. PMID: 37536571 Review. - The staphylococcal surface-glycopolymer wall teichoic acid (WTA) is crucial for complement activation and immunological defense against Staphylococcus aureus infection.
Kurokawa K, Takahashi K, Lee BL. Kurokawa K, et al. Immunobiology. 2016 Oct;221(10):1091-101. doi: 10.1016/j.imbio.2016.06.003. Epub 2016 Jun 15. Immunobiology. 2016. PMID: 27424796 Review.
Cited by
- Interactions of the cell-wall glycopolymers of lactic acid bacteria with their bacteriophages.
Chapot-Chartier MP. Chapot-Chartier MP. Front Microbiol. 2014 May 22;5:236. doi: 10.3389/fmicb.2014.00236. eCollection 2014. Front Microbiol. 2014. PMID: 24904550 Free PMC article. Review. - Lipoproteins Contribute to the Anti-inflammatory Capacity of Lactobacillus plantarum WCFS1.
Lee IC, van Swam II, Boeren S, Vervoort J, Meijerink M, Taverne N, Starrenburg M, Bron PA, Kleerebezem M. Lee IC, et al. Front Microbiol. 2020 Jul 29;11:1822. doi: 10.3389/fmicb.2020.01822. eCollection 2020. Front Microbiol. 2020. PMID: 32849426 Free PMC article. - The role of cell surface architecture of lactobacilli in host-microbe interactions in the gastrointestinal tract.
Sengupta R, Altermann E, Anderson RC, McNabb WC, Moughan PJ, Roy NC. Sengupta R, et al. Mediators Inflamm. 2013;2013:237921. doi: 10.1155/2013/237921. Epub 2013 Mar 13. Mediators Inflamm. 2013. PMID: 23576850 Free PMC article. Review. - Postbiotics as the key mediators of the gut microbiota-host interactions.
Ozma MA, Abbasi A, Akrami S, Lahouty M, Shahbazi N, Ganbarov K, Pagliano P, Sabahi S, Köse Ş, Yousefi M, Dao S, Asgharzadeh M, Hosseini H, Kafil HS. Ozma MA, et al. Infez Med. 2022 Jun 1;30(2):180-193. doi: 10.53854/liim-3002-3. eCollection 2022. Infez Med. 2022. PMID: 35693065 Free PMC article. Review. - The sweet tooth of bacteria: common themes in bacterial glycoconjugates.
Tytgat HL, Lebeer S. Tytgat HL, et al. Microbiol Mol Biol Rev. 2014 Sep;78(3):372-417. doi: 10.1128/MMBR.00007-14. Microbiol Mol Biol Rev. 2014. PMID: 25184559 Free PMC article. Review.
References
- Bron PA, van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol. 2012;10:66–78. - PubMed
- Xia G, Kohler T, Peschel A. The wall teichoic acid and lipoteichoic acid polymers ofStaphylococcus aureus. Int J Med Microbiol. 2010;300(2–3):148–154. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases