The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr Virus - PubMed (original) (raw)
The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr Virus
Chung-Pei Lee et al. PLoS Pathog. 2012 Sep.
Abstract
The cellular endosomal sorting complex required for transport (ESCRT) machinery participates in membrane scission and cytoplasmic budding of many RNA viruses. Here, we found that expression of dominant negative ESCRT proteins caused a blockade of Epstein-Barr virus (EBV) release and retention of viral BFRF1 at the nuclear envelope. The ESCRT adaptor protein Alix was redistributed and partially colocalized with BFRF1 at the nuclear rim of virus replicating cells. Following transient transfection, BFRF1 associated with ESCRT proteins, reorganized the nuclear membrane and induced perinuclear vesicle formation. Multiple domains within BFRF1 mediated vesicle formation and Alix recruitment, whereas both Bro and PRR domains of Alix interacted with BFRF1. Inhibition of ESCRT machinery abolished BFRF1-induced vesicle formation, leading to the accumulation of viral DNA and capsid proteins in the nucleus of EBV-replicating cells. Overall, data here suggest that BFRF1 recruits the ESCRT components to modulate nuclear envelope for the nuclear egress of EBV.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
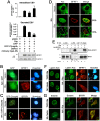
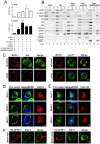
Figure 1. The ESCRT machinery contributes to EBV maturation and is redistributed to nucleus-associated compartments in EBV-reactivated NA cells.
(A) EBV-positive NA cells were transfected with plasmids expressing GFP-Chmp4b, GFP (the GFP vector), mCherry-Vps4-DN or DsRed (the mCherry related vector) together with a plasmid expressing Rta or pSG5 vector control. The viral DNA content in the transfected cells and culture supernatant was harvested at 96 h post induction and subjected to qPCR analysis targeting the EBV DNA _Bam_HI W fragment. Results are means ± standard deviations from two separate transfections. Data are representatives of two independent experiments. The statistically significant differences between the Rta expression alone and coexpression of Rta and GFP-Chmp4b or mCherry-Vps4-DN are calculated by the paired Student's t-test and indicated at the top of the bars. *, P<0.05; **, P<0.01. The expressed EBV BFRF1 and cellular lamin A proteins also were detected by immunoblotting using anti-BFRF1 (E10) anti-lamin A/C. Expression of GFP-Chmp4b or Vps4-DN reduced the virion secretion. (B and C) To investigate the GFP-Chmp4b localization in EBV reactivated cells, slide-cultured NA cells were transfected with plasmids expressing GFP or GFP-Chmp4b, together with Rta expressing plasmid. At 48 h post transfection, cells were fixed by 4% paraformaldehyde and immuno-stained for BFRF1 (red). Cellular DNA was detected by Hoechst 33258 staining. (D) To detect Alix protein in EBV reactivated cells, slide-cultured NA cells were transfected with Rta expressing or vector plasmid. At 48 h post transfection, cells were then fixed, stained for Alix (green) and BFRF1 (red) and observed using confocal microscopy. The cell margin in b was detected by MetaMorph software and is indicated by a white outline. Two major types of BFRF1 distribution were observed in virus- reactivated cells: a, nuclear rim distribution (∼65%) and b, nuclear rim with cytoplasmic punctate distribution (∼35%). (E) Lysates from Rta-transfected NA cells were immunoprecipitated with antibody against Alix, HA or GST antibody as a negative control. The immunocomplexes were then resolved by 10% SDS-PAGE and immunoblotted with antibodies against HA or Alix. The coimmunoprecipitated proteins are indicated by “*”. (F and G) To observe the nuclear envelope structure in EBV-reactivated cells, NA or NPC-TW01 cells were transfected with a plasmid expressing Rta to induce virus lytic replication or vector plasmid. At 72 h post transfection, cells were fixed and immuno-stained for lamin A/C (green) and BFRF1 (red), or emerin (green) and BFRF1 (red) and stained with cellular DNA. Approximately 50∼100 cells with BFRF1 expression were counted for the appearance of cytoplasmic vesicle associated lamin A/C or emerin in each set of transfection, and all the experiments were repeated independently at least twice.
Figure 2. EBV BFRF1 reorganizes the nuclear membrane and induces vesicle formation from the altered membrane.
(A to D) Slide-cultured HeLa cells were transfected with a plasmid expressing HA-BFRF1, Flag-BFLF2 or control vector for 24 h. The cells were fixed with 4% paraformaldehyde and stained for HA (A and B, red), Flag (C and D, red), lamin A/C (A and C, green) or emerin (B and D, green), and DNA. Approximately 50∼100 HA-BFRF1 expressing cells were counted to detect the cytoplasmic vesicle associated lamin (A and C) or emerin (B and D) in each set of transfections and all the experiments were repeated independently at least twice. (E) To investigate the effects of BFRF1 on nucleoporins distribution, HeLa cells were transfected with a plasmid expressing HA-BFRF1 or control vector for 24 h, stained for HA-BFRF1 (red) or FG-repeat containing nucleoporins (mAb414, green) and DNA, and analyzed by confocal microscopy. (F) HeLa cells were cotransfected with plasmid expressing HA-BFRF1 and Flag-BFLF2 for 24 h, fixed, stained for HA, Flag and DNA, and analyzed by confocal microscopy.
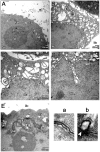
Figure 3. TEM analysis of cells with EBV BFRF1 expression.
HeLa cells were transfected with control vector (A) or plasmid expressing HA-BFRF1 (B to E). At 24 h post transfection, cells were fixed and processed for TEM as described in Materials and Methods. More than 30 cells were observed in each set and representative nuclear and cytoplasmic regions of transfected cells are shown. Images were taken at a final magnification of ×12,000 (A and B) or ×20,000 (C to E). (A and B) Expression of HA-BFRF1 induced multiple irregular shape vesicles in cytoplasm (Cy) and altered nuclear membrane structure (NM). (C) Irregular shape vesicles with single or multiple-layer membranes (arrowhead) were observed in cells transfected with HA-BFRF1. (D) A wavy NM was observed in a nuclear cave structure of HA-BFRF1 transfected cell. (E) An untypical multilayered NM (a) and a budding nuclear membrane (b) were shown at higher magnification in the inset, respectively. “*”, indistinct margin of NM. Cy, cytoplasm; Nu, nucleus; Mt, mitochondria; Ex, extracellular space.
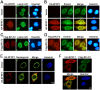
Figure 4. Cellular ESCRT components contribute to BFRF1-mediated vesicle formation.
(A) Slide-cultured HeLa cells were transfected with plasmids expressing wild-type GFP-Vps4A and HA-BFRF1 for 24 h, and stained for HA (red) and DNA. (B) HeLa cells were transfected with plasmids expressing Vps4-DN and HA-BFRF1 for 24 h and stained for HA (red) and DNA. The original color of HA (green) and mCherry (red) were converted to red and green to facilitate interpretation of the data. Nuclear membrane associated HA-BFRF1 and Vps4-DN colocalized at nuclear-associated clusters without cytoplasmic vesicle structure (yellow, Merge). (C) Slide-cultured HeLa cells were transfected with plasmid expressing HA-BFRF1. At 24 h post transfection, cells were fixed and immuno-stained for HA (red) and endogenous Alix (green). Expression of BFRF1 redistributed Alix to nuclear rim. (D) Lysates from HeLa cells transfected with vector or plasmid expressing HA-BFRF1, GFP-Chmp4b or mCherry-Vps4-DN were immunoprecipitated with antibody against GFP, HA or GST control antibody. The immunocomplexes were then resolved by 10% SDS-PAGE and immunoblotted with antibodies against HA, GFP, Alix or Vps4. BFRF1 associated with multiple ESCRT proteins. (E) To check the knockdown efficiency of siRNA treatment, HeLa cells were transfected with Alix or control (Ctl) siRNA for 48, 72 or 96 h, harvested and subjected to immunoblotting analysis using anti-Alix or anti-GAPDH antibody. (F) HeLa cells were transfected with Alix (siAlix) or control (siControl) siRNA for 48 h and cotransfected with siRNAs and plasmid expressing HA-BFRF1 for additional 24 h. The transfected cells were then fixed and immuno-stained for HA (red) and Alix (green). Knockdown of Alix expression abolished BFRF1 induced vesicle formation.
Figure 5. Functional domain mapping of BFRF1 and Alix required for the vesicle formation and molecular interaction.
(A) Schematic representation of HA-tagged BFRF1 mutants. Putative late domain, BFRF2 interacting domain, EBV specific region and transmembrane domain were predicted by MUSCLE and ClustalW2 multiple alignment (
http://www.ebi.ac.uk/Tools/msa/muscle/and/clustalw2/
), and DAS protein structure modeling (
http://www.sbc.su.se/\~miklos/DAS/
) programs. A series of BFRF1 deletion mutants, including ΔLD1, ΔLD2, ΔID, ΔESR and ΔTM, were generated as indicated. The relative Alix binding ability was evaluated in coimmunoprecipitated protein signals, and summarized in the right-hand column. (B) Slide-cultured HeLa cells were transfected with HA-BFRF1 wild-type (WT), ΔLD1, ΔLD2, ΔID, ΔESR or ΔTM-expressing plasmids, fixed by 4% paraformaldehyde, stained for HA (red), Alix (green) and DNA and observed in confocal microscopy. (C) Lysates from HeLa cells transfected with HA-pSG5 vector (VC) or plasmid expressing WT, ΔLD1, ΔLD2, ΔID, ΔESR or ΔTM of HA-BFRF1 were immunoprecipitated with antibody against Alix (upper panel), HA (lower panel) or anti-GST control antibody. The immunocomplexes were then resolved by 10% SDS-PAGE and immunoblotted with antibodies against Alix and HA. IgL, immunoglobulin light chain. (D) Schematic representation of Flag-tagged Alix mutants. Alix functional domain mutants, including Alix_Bro, Alix_V and Alix_PRR were used as indicated. WT, wild type; Bro, Bro1 domain; V, “V” domain; PRR, proline-rich region. (E) WT, Bro and PRR fragments of Alix were coimmunoprecipitated with HA-BFRF1. Lysates from HeLa cells transfected with vector or plasmid expressing HA-BFRF1 coupled with Flag-Alix, Flag-Alix_Bro, Flag-Alix_V or Flag-Alix_PRR. HA-BFRF1 were immunoprecipitated with antibody against HA or GST. The immunocomplexes were then resolved by 10% SDS-PAGE and immunoblotted with antibodies against Flag, Alix, or HA.
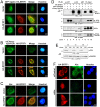
Figure 6. Expression of Alix siRNA, dominant negative Vps4 or BFRF1ΔESR accumulates viral genome and capsid proteins in the nucleus.
(A and B) To observe the effect of Alix siRNA, BFRF1ΔESR or Vps-DN expression on the viral genome and distribution of capsid proteins, NA cells were transfected with plasmid expressing Rta or vector pSG5 and siAlix or siControl for 48 h and followed by another transfection of siRNA or plasmid expressing mCherry-Vps4-DN, BFRF1ΔESR, or related control vector, and incubated for additional 48 h. The subcellular fractionation was then performed as described in Materials and Methods. DNA and protein were extracted individually from transfected cells into total (T, or intracellular), nuclear (N) and cytosol (C) fractions. The extracted DNA in the total and nuclear fraction was then subjected to qPCR analysis targeting the EBV DNA _Bam_HI W fragment and the relative folds of viral genome in intracellular or nuclear compartment of transfected cells are indicated in (A). Results are means ± standard deviations from two separate transfections. Data are representatives of two independent experiments. The statistically significant differences between Rta expression alone and coexpression of Rta and siAlix, mCherry-Vps4-DN or HA-BFRF1ΔESR were calculated by the paired Student's t-test and are indicated at the top of the bars. *, P<0.05; ***, P<0.001. The protein expression in the various factions is shown in (B). PARP and α-Tubulin serves as nuclear and cytosolic markers, respectively. (C) NA cells were transfected with plasmid expressing Rta together with siAlix or siControl for 48 h and followed by another transfection of siRNA for additional 24 h. The cells were fixed by 4% paraformaldehyde and immune-stained for Alix, BFRF1 and capsid protein BcLF1, respectively. The margin of the cells was detected by MetaMorph software and is indicated by a white outline. Treatment with siAlix induced the accumulation of BcLF1 in the nucleus. (D and E) To observe the localization of nucleocapsids, lytic NA cells with DsRed control (red) or mCherry-Vps4-DN (red) expression were fixed at 72 h post transfection, immuno-stained for the subcellular distribution of major viral capsid components BcLF1, BORF1 or BDLF1 (green) with anti-BcLF1 L2, anti-BORF1 or anti-BDLF1 antibody, and observed by confocal microscopy. Cellular DNA was detected by Hoechst 33258 and merged with capsid protein staining. Expression of Vps4-DN promoted the intra-nuclear accumulation of capsid components BcLF1, BORF1 and BDLF1 (overlaid with blue DNA signals). (F) NA cells were transfected with plasmid expressing Rta for 48 h followed by transfection of HA-BFRF1 or BFRF1ΔESR plasmid for an additional 24 h. Transfected cells were fixed, immuno-stained for viral major capsid component BcLF1 (green) and HA (red) and observed by confocal microscopy. BFRF1ΔESR of expression reduced the BcLF1 detection in cytoplasm of transfected cells.
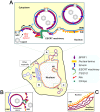
Figure 7. A hypothetical model of the interaction among EBV BFRF1 and cellular ESCRT components in vesicle formation and modulation of the nuclear membrane.
After EBV reactivation, membrane anchoring BFRF1 targets to the nucleus associated membranes. BFRF1 potentially regulates the membrane through protein oligomerization and then recruits Alix and, sequentially, other ESCRT components to derive the vesicles from the double layer nuclear envelope (A), outer nuclear membrane (ONM) or from inner nuclear membrane (INM) (B). These nuclear membrane-derived vesicles may contain some nucleoporins and other INM proteins, such as lamin A/C or emerin. Cooperating with other viral factors, such as BFLF2, the BFRF1-mediated vesicles may further recruit and pack some nucleocapsids for the subsequent nuclear egress. Additionally, the expression of BFRF1 induces the formation of multilayered cisternal nuclear membranes (C), which may provide more membranous structure for viral budding. ER, endoplasmic reticulum. NPC, nuclear pore complex.
Similar articles
- The Ubiquitin Ligase Itch and Ubiquitination Regulate BFRF1-Mediated Nuclear Envelope Modification for Epstein-Barr Virus Maturation.
Lee CP, Liu GT, Kung HN, Liu PT, Liao YT, Chow LP, Chang LS, Chang YH, Chang CW, Shu WC, Angers A, Farina A, Lin SF, Tsai CH, Bouamr F, Chen MR. Lee CP, et al. J Virol. 2016 Sep 29;90(20):8994-9007. doi: 10.1128/JVI.01235-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27466427 Free PMC article. - The Novel Nuclear Targeting and BFRF1-Interacting Domains of BFLF2 Are Essential for Efficient Epstein-Barr Virus Virion Release.
Dai YC, Liao YT, Juan YT, Cheng YY, Su MT, Su YZ, Liu HC, Tsai CH, Lee CP, Chen MR. Dai YC, et al. J Virol. 2020 Jan 17;94(3):e01498-19. doi: 10.1128/JVI.01498-19. Print 2020 Jan 17. J Virol. 2020. PMID: 31694953 Free PMC article. - BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress.
Farina A, Feederle R, Raffa S, Gonnella R, Santarelli R, Frati L, Angeloni A, Torrisi MR, Faggioni A, Delecluse HJ. Farina A, et al. J Virol. 2005 Mar;79(6):3703-12. doi: 10.1128/JVI.79.6.3703-3712.2005. J Virol. 2005. PMID: 15731264 Free PMC article. - Seeking Closure: How Do Herpesviruses Recruit the Cellular ESCRT Apparatus?
Barnes J, Wilson DW. Barnes J, et al. J Virol. 2019 Jun 14;93(13):e00392-19. doi: 10.1128/JVI.00392-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996099 Free PMC article. Review. - Conquering the Nuclear Envelope Barriers by EBV Lytic Replication.
Lee CP, Chen MR. Lee CP, et al. Viruses. 2021 Apr 18;13(4):702. doi: 10.3390/v13040702. Viruses. 2021. PMID: 33919628 Free PMC article. Review.
Cited by
- The Ubiquitin Ligase Itch and Ubiquitination Regulate BFRF1-Mediated Nuclear Envelope Modification for Epstein-Barr Virus Maturation.
Lee CP, Liu GT, Kung HN, Liu PT, Liao YT, Chow LP, Chang LS, Chang YH, Chang CW, Shu WC, Angers A, Farina A, Lin SF, Tsai CH, Bouamr F, Chen MR. Lee CP, et al. J Virol. 2016 Sep 29;90(20):8994-9007. doi: 10.1128/JVI.01235-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27466427 Free PMC article. - The many functions of ESCRTs.
Vietri M, Radulovic M, Stenmark H. Vietri M, et al. Nat Rev Mol Cell Biol. 2020 Jan;21(1):25-42. doi: 10.1038/s41580-019-0177-4. Epub 2019 Nov 8. Nat Rev Mol Cell Biol. 2020. PMID: 31705132 Review. - Vitamin D3/VDR inhibits inflammation through NF-κB pathway accompanied by resisting apoptosis and inducing autophagy in abalone Haliotis discus hannai.
Huang D, Guo Y, Li X, Pan M, Liu J, Zhang W, Mai K. Huang D, et al. Cell Biol Toxicol. 2023 Jun;39(3):885-906. doi: 10.1007/s10565-021-09647-4. Epub 2021 Oct 12. Cell Biol Toxicol. 2023. PMID: 34637036 - Atg39 links and deforms the outer and inner nuclear membranes in selective autophagy of the nucleus.
Mochida K, Otani T, Katsumata Y, Kirisako H, Kakuta C, Kotani T, Nakatogawa H. Mochida K, et al. J Cell Biol. 2022 Feb 7;221(2):e202103178. doi: 10.1083/jcb.202103178. Epub 2022 Jan 21. J Cell Biol. 2022. PMID: 35061008 Free PMC article. - Analysis of RNA-Seq datasets reveals enrichment of tissue-specific splice variants for nuclear envelope proteins.
Capitanchik C, Dixon CR, Swanson SK, Florens L, Kerr ARW, Schirmer EC. Capitanchik C, et al. Nucleus. 2018;9(1):410-430. doi: 10.1080/19491034.2018.1469351. Nucleus. 2018. PMID: 29912636 Free PMC article.
References
- Raiborg C, Stenmark H (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458: 445–452. - PubMed
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG (2003) AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114: 689–699. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous