Hepatic lipid accumulation alters global histone h3 lysine 9 and 4 trimethylation in the peroxisome proliferator-activated receptor alpha network - PubMed (original) (raw)
Hepatic lipid accumulation alters global histone h3 lysine 9 and 4 trimethylation in the peroxisome proliferator-activated receptor alpha network
Hee-Jin Jun et al. PLoS One. 2012.
Erratum in
- Correction: Hepatic Lipid Accumulation Alters Global Histone H3 Lysine 9 and 4 Trimethylation in the Peroxisome Proliferator-Activated Receptor Alpha Network.
Jun HJ, Kim J, Hoang MH, Lee SJ. Jun HJ, et al. PLoS One. 2012 Nov 30;7(11):10.1371/annotation/eff6e471-306a-41bd-88e3-13857af094af. doi: 10.1371/annotation/eff6e471-306a-41bd-88e3-13857af094af. eCollection 2012. PLoS One. 2012. PMID: 29364954 Free PMC article.
Abstract
Recent data suggest that the etiology of several metabolic diseases is closely associated with transcriptome alteration by aberrant histone methylation. We performed DNA microarray and ChIP-on-chip analyses to examine transcriptome profiling and trimethylation alterations to identify the genomic signature of nonalcoholic fatty liver disease (NAFLD), the most common form of chronic liver disease. Transcriptome analysis showed that steatotic livers in high-fat diet-fed apolipoprotein E2 mice significantly altered the expression of approximately 70% of total genes compared with normal diet-fed control livers, suggesting that hepatic lipid accumulation induces dramatic alterations in gene expression in vivo. Also, pathway analysis suggested that genes encoding chromatin-remodeling enzymes, such as jumonji C-domain-containing histone demethylases that regulate histone H3K9 and H3K4 trimethylation (H3K9me3, H3K4me3), were significantly altered in steatotic livers. Thus, we further investigated the global H3K9me3 and H3K4me3 status in lipid-accumulated mouse primary hepatocytes by ChIP-on-chip analysis. Results showed that hepatic lipid accumulation induced aberrant H3K9me3 and H3K4me3 status in peroxisome proliferator-activated receptor alpha and hepatic lipid catabolism network genes, reducing their mRNA expression compared with non-treated control hepatocytes. This study provides the first evidence that epigenetic regulation by H3K9me3 and H3K4me3 in hepatocytes may be involved in hepatic steatosis and the pathogenesis of NAFLD. Thus, control of H3K9me3 and H3K4me3 represents a potential novel NAFLD prevention and treatment strategy.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
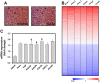
Figure 1. Transcriptome profile of the steatotic livers of high-fat diet-fed hAPOE2 mice determined by oligonucleotide microarray analysis.
(A) H&E staining of the livers of normal diet- and high-fat diet-fed hAPOE2 mice (original magnification, ×400). (B) Heat map of the transcriptome profile of the steatotic livers of high-fat diet-fed hAPOE2 mice. Columns represent individual arrays and rows indicate gene expression profiles. Red, blue, and white indicate upregulated, downregulated, and unaltered genes, respectively (p<0.05, n = 6). (C) mRNA expression of genes encoding epigenetic modifiers in the steatotic livers of high-fat diet-fed hAPOE2 mice (p<0.05).
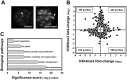
Figure 2. Genome-wide H3K9me3 and H3K4me3 variations in lipid-accumulated mouse primary hepatocytes determined by ChIP-on-chip analysis.
(A) BODIPY-labeled lipid droplets in non- and palmitate plus oleate-treated mouse primary hepatocytes. (B) Expression pattern of H3K9me3 and H3K4me3 targets (fold change ≥1.5 in at least one histone trimethylation status, p<0.05, n = 2 for each histone status, H3K9me3 and H3K4me3) in lipid-accumulated hepatocytes. (C) Biological pathways affected by H3K9me3 and H3K4me3 targets in response to lipid accumulation in hepatocytes (p<0.05).
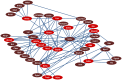
Figure 3. Effect of H3K4me3 and H3K9me3 on the PPARα-network in lipid-accumulated mouse primary hepatocytes.
Lipid metabolism-associated H3K9me3 and H3K4me3 targets (p<0.05) were selected based on Gene Ontology annotation, and their biological relationship was analyzed using the PubGene database. Red indicates H3K9me3 and H3K4me3 targets detected by ChIP-on-chip analysis and dark red represents those genes possessing a potential biological relationship with targets detected by ChIP-on-chip analysis based on previous reports.
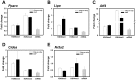
Figure 4. ChIP analysis of mRNA levels of H3K9me3 and H3K4me3 and selected Pparα network genes (n = 3).
Similar articles
- MacroH2A1 isoforms are associated with epigenetic markers for activation of lipogenic genes in fat-induced steatosis.
Podrini C, Koffas A, Chokshi S, Vinciguerra M, Lelliott CJ, White JK, Adissu HA, Williams R, Greco A. Podrini C, et al. FASEB J. 2015 May;29(5):1676-87. doi: 10.1096/fj.14-262717. Epub 2014 Dec 19. FASEB J. 2015. PMID: 25526730 - Obesity increases histone H3 lysine 9 and 18 acetylation at Tnfa and Ccl2 genes in mouse liver.
Mikula M, Majewska A, Ledwon JK, Dzwonek A, Ostrowski J. Mikula M, et al. Int J Mol Med. 2014 Dec;34(6):1647-54. doi: 10.3892/ijmm.2014.1958. Epub 2014 Oct 3. Int J Mol Med. 2014. PMID: 25319795 - Histone H3K9 demethylase JMJD2B induces hepatic steatosis through upregulation of PPARγ2.
Kim JH, Jung DY, Nagappan A, Jung MH. Kim JH, et al. Sci Rep. 2018 Sep 13;8(1):13734. doi: 10.1038/s41598-018-31953-x. Sci Rep. 2018. PMID: 30214048 Free PMC article. - Genetic and epigenetic variants influencing the development of nonalcoholic fatty liver disease.
Li YY. Li YY. World J Gastroenterol. 2012 Dec 7;18(45):6546-51. doi: 10.3748/wjg.v18.i45.6546. World J Gastroenterol. 2012. PMID: 23236228 Free PMC article. Review. - Reactive oxygen and nitrogen species in steatotic hepatocytes: a molecular perspective on the pathophysiology of ischemia-reperfusion injury in the fatty liver.
Reiniers MJ, van Golen RF, van Gulik TM, Heger M. Reiniers MJ, et al. Antioxid Redox Signal. 2014 Sep 1;21(7):1119-42. doi: 10.1089/ars.2013.5486. Epub 2014 Feb 19. Antioxid Redox Signal. 2014. PMID: 24294945 Free PMC article. Review.
Cited by
- Dietary Patterns Influence Target Gene Expression through Emerging Epigenetic Mechanisms in Nonalcoholic Fatty Liver Disease.
Zaiou M, Amrani R, Rihn B, Hajri T. Zaiou M, et al. Biomedicines. 2021 Sep 18;9(9):1256. doi: 10.3390/biomedicines9091256. Biomedicines. 2021. PMID: 34572442 Free PMC article. Review. - Non-alcoholic fatty liver disease--a chronic disease of the 21st century.
Metrakos P, Nilsson T. Metrakos P, et al. J Biomed Res. 2018 Sep 29;32(5):327-335. doi: 10.7555/JBR.31.20160153. J Biomed Res. 2018. PMID: 28550272 Free PMC article. - Strategies, models and biomarkers in experimental non-alcoholic fatty liver disease research.
Willebrords J, Pereira IV, Maes M, Crespo Yanguas S, Colle I, Van Den Bossche B, Da Silva TC, de Oliveira CP, Andraus W, Alves VA, Cogliati B, Vinken M. Willebrords J, et al. Prog Lipid Res. 2015 Jul;59:106-25. doi: 10.1016/j.plipres.2015.05.002. Epub 2015 Jun 11. Prog Lipid Res. 2015. PMID: 26073454 Free PMC article. Review. - The use of transcriptomics to unveil the role of nutrients in Mammalian liver.
Osada J. Osada J. ISRN Nutr. 2013 Aug 28;2013:403792. doi: 10.5402/2013/403792. eCollection 2013. ISRN Nutr. 2013. PMID: 24967258 Free PMC article. Review. - Amelioration of diet-induced steatohepatitis in mice following combined therapy with ASO-Fsp27 and fenofibrate.
Rajamoorthi A, Arias N, Basta J, Lee RG, Baldán Á. Rajamoorthi A, et al. J Lipid Res. 2017 Nov;58(11):2127-2138. doi: 10.1194/jlr.M077941. Epub 2017 Sep 5. J Lipid Res. 2017. PMID: 28874443 Free PMC article.
References
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080. - PubMed
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45. - PubMed
- Margueron R, Trojer P, Reinberg D (2005) The key to development: interpreting the histone code? Curr Opin Genet Dev 15: 163–176. - PubMed
- Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15: 2343–2360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases