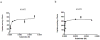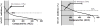Potent inhibitors of human organic anion transporters 1 and 3 from clinical drug libraries: discovery and molecular characterization - PubMed (original) (raw)
. 2012 Nov 5;9(11):3340-6.
doi: 10.1021/mp300365t. Epub 2012 Sep 25.
Affiliations
- PMID: 22973893
- PMCID: PMC3490050
- DOI: 10.1021/mp300365t
Potent inhibitors of human organic anion transporters 1 and 3 from clinical drug libraries: discovery and molecular characterization
Peng Duan et al. Mol Pharm. 2012.
Abstract
Transporter-mediated drug-drug interactions in the kidney dramatically influence the pharmacokinetics and other clinical effects of drugs. Human organic anion transporters 1 (hOAT1) and 3 (hOAT3) are the major transporters in the basolateral membrane of kidney proximal tubules, mediating the rate-limiting step in the elimination of a broad spectrum of drugs. In the present study, we screened two clinical drug libraries against hOAT1 and hOAT3. Of the 727 compounds screened, 92 compounds inhibited hOAT1 and 262 compounds inhibited hOAT3. When prioritized based on the peak unbound plasma concentrations of these compounds, three inhibitors for hOAT1 and seven inhibitors for hOAT3 were subsequently identified with high inhibitory potency (>95%). Computational analyses revealed that inhibitors and noninhibitors can be differentiated from each other on the basis of several physicochemical features, including number of hydrogen-bond donors, number of rotatable bonds, and topological polar surface area (TPSA) for hOAT1; and molecular weight, number of hydrogen-bond donors and acceptors, TPSA, partition coefficient (log P(7.4)), and polarizability for hOAT3. Pharmacophore modeling identified two common structural features associated with inhibitors for hOAT1 and hOAT3, viz., an anionic hydrogen-bond acceptor atom, and an aromatic center separated by ∼5.7 Å. Such model provides mechanistic insights for predicting new OAT inhibitors.
Figures
Fig. 1. Concentration dependence of 6-CF uptake in hOAT1- and hOAT3- expressing cells
a. The hOAT1-mediated specific uptake was calculated by subtracting the nonspecific uptake in control cells from that in the cells expressing hOAT1. b. The hOAT3-mediated specific uptake was calculated by subtracting the nonspecific uptake in control cells from that in the cells expressing hOAT3.
Fig. 2. Inhibitors of hOAT1 and hOAT3 identified in a screen of 727 clinically tested drugs
a. Overview of the results from the screening of hOAT1 inhibition. The 92 compounds (at 50μM) resulting in at least 50% decreased uptake of 6-CF were classified as inhibitors (shaded in light gray and under the curve). Data were presented as the mean ± SD (samples in triplicate from one experiment). b. Overview of the results from the screening of hOAT3 inhibition. The 262 compounds (at 20μM) resulting in at least 50% decreased uptake of 6-CF were classified as inhibitors (shaded in light gray and under the curve). Data were presented as the mean ± SD (samples in triplicate from one experiment).
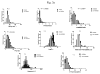
Fig. 3. Structural features of hOAT1 and hOAT3 inhibitors
a. Distribution of key physicochemical properties of hOAT1 inhibitors. b. Distribution of key physicochemical properties of hOAT3 inhibitors. The calculated descriptors (viz., TPSA, molecular weight, volume, number of rotatable bonds, Log P7.4, molecular polarizability, number of hydrogen-bond donors and hydrogen-bond acceptors) were calculated using Chemaxon JChem package (ChemAxon, USA). The frequency distribution analysis was conducted between inhibitors and non-inhibitors by Graphpad prism 5.0. *P < 0.05 was considered significantly different, and n.s. stood for no significant difference.
Fig. 3. Structural features of hOAT1 and hOAT3 inhibitors
a. Distribution of key physicochemical properties of hOAT1 inhibitors. b. Distribution of key physicochemical properties of hOAT3 inhibitors. The calculated descriptors (viz., TPSA, molecular weight, volume, number of rotatable bonds, Log P7.4, molecular polarizability, number of hydrogen-bond donors and hydrogen-bond acceptors) were calculated using Chemaxon JChem package (ChemAxon, USA). The frequency distribution analysis was conducted between inhibitors and non-inhibitors by Graphpad prism 5.0. *P < 0.05 was considered significantly different, and n.s. stood for no significant difference.
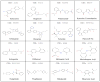
Fig. 4
Structures of inhibitors used in pharmacophore analysis.
Fig. 5. Pharmacophore model of hOAT1 and hOAT3 inhibitors
Two representative OAT inhibitors were mapped to the proposed pharmacophore model. The anionic and hydrogen-bond acceptor atom is displayed as cyan sphere and labeled F1:Ani &Acc, while the aromatic center is represented as a yellow sphere and labeled F2:Aro. Carbon atoms in the compound (CID: 84003) is colored in magenta and those in the compound (CID: 60560) is shown in gray. The other atoms in the compounds are colored by atom type (nitrogen: blue; oxygen: red; sulfur: yellow).
Similar articles
- Effects of phenylpropanoids on human organic anion transporters hOAT1 and hOAT3.
Kawasaki T, Takeichi Y, Tomita M, Uwai Y, Epifano F, Fiorito S, Taddeo VA, Genovese S, Nabekura T. Kawasaki T, et al. Biochem Biophys Res Commun. 2017 Aug 5;489(4):375-380. doi: 10.1016/j.bbrc.2017.05.121. Epub 2017 May 27. Biochem Biophys Res Commun. 2017. PMID: 28559139 - Interaction and transport characteristics of mycophenolic acid and its glucuronide via human organic anion transporters hOAT1 and hOAT3.
Uwai Y, Motohashi H, Tsuji Y, Ueo H, Katsura T, Inui K. Uwai Y, et al. Biochem Pharmacol. 2007 Jun 30;74(1):161-8. doi: 10.1016/j.bcp.2007.03.024. Epub 2007 Mar 30. Biochem Pharmacol. 2007. PMID: 17462604 - Proton pump inhibitors inhibit methotrexate transport by renal basolateral organic anion transporter hOAT3.
Chioukh R, Noel-Hudson MS, Ribes S, Fournier N, Becquemont L, Verstuyft C. Chioukh R, et al. Drug Metab Dispos. 2014 Dec;42(12):2041-8. doi: 10.1124/dmd.114.058529. Epub 2014 Sep 19. Drug Metab Dispos. 2014. PMID: 25239859 - Characterization of ochratoxin A transport by human organic anion transporters.
Jung KY, Takeda M, Kim DK, Tojo A, Narikawa S, Yoo BS, Hosoyamada M, Cha SH, Sekine T, Endou H. Jung KY, et al. Life Sci. 2001 Sep 21;69(18):2123-35. doi: 10.1016/s0024-3205(01)01296-6. Life Sci. 2001. PMID: 11669456 - Can we Predict Drug Excretion into Saliva? A Systematic Review and Analysis of Physicochemical Properties.
Nguyen TA, Chen RH, Hawkins BA, Hibbs DE, Kim HY, Wheate NJ, Groundwater PW, Stocker SL, Alffenaar JC. Nguyen TA, et al. Clin Pharmacokinet. 2024 Aug;63(8):1067-1087. doi: 10.1007/s40262-024-01398-9. Epub 2024 Jul 15. Clin Pharmacokinet. 2024. PMID: 39008243 Free PMC article.
Cited by
- Inhibitory effects of chemotherapeutics on human organic anion transporter hOAT4.
Toh MF, Suh W, Wang H, Zhou P, Hu L, You G. Toh MF, et al. Int J Biochem Mol Biol. 2016 Jun 1;7(1):11-8. eCollection 2016. Int J Biochem Mol Biol. 2016. PMID: 27335682 Free PMC article. - Deuterated Indocyanine Green (ICG) with Extended Aqueous Storage Shelf-Life: Chemical and Clinical Implications.
Li DH, Smith BD. Li DH, et al. Chemistry. 2021 Oct 19;27(58):14535-14542. doi: 10.1002/chem.202102816. Epub 2021 Sep 24. Chemistry. 2021. PMID: 34403531 Free PMC article. - Inhibition of Human Drug Transporter Activities by the Pyrethroid Pesticides Allethrin and Tetramethrin.
Chedik L, Bruyere A, Le Vee M, Stieger B, Denizot C, Parmentier Y, Potin S, Fardel O. Chedik L, et al. PLoS One. 2017 Jan 18;12(1):e0169480. doi: 10.1371/journal.pone.0169480. eCollection 2017. PLoS One. 2017. PMID: 28099443 Free PMC article. - Position-related renal perfusion disturbances as a possible underestimated mechanism in patients with resistant hypertension: a case vignette.
Schiefer J, Amthauer H, Genseke P, Mertens PR, Chatzikyrkou C. Schiefer J, et al. Int Urol Nephrol. 2017 Oct;49(10):1823-1833. doi: 10.1007/s11255-017-1656-1. Epub 2017 Jul 11. Int Urol Nephrol. 2017. PMID: 28699016 - Posttranslational Regulation of Organic Anion Transporters by Ubiquitination: Known and Novel.
Xu D, Wang H, You G. Xu D, et al. Med Res Rev. 2016 Sep;36(5):964-79. doi: 10.1002/med.21397. Epub 2016 Jun 12. Med Res Rev. 2016. PMID: 27291023 Free PMC article. Review.
References
- You G. Structure, function, and regulation of renal organic anion transporters. Medicinal research reviews. 2002;22(6):602–16. - PubMed
- Dantzler WH, Wright SH. The molecular and cellular physiology of basolateral organic anion transport in mammalian renal tubules. Biochimica et biophysica acta. 2003;1618(2):185–93. - PubMed
- VanWert AL, Gionfriddo MR, Sweet DH. Organic anion transporters: discovery, pharmacology, regulation and roles in pathophysiology. Biopharmaceutics & drug disposition. 2010;31(1):1–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM097000/GM/NIGMS NIH HHS/United States
- R01-GM079123/GM/NIGMS NIH HHS/United States
- R01 GM079123/GM/NIGMS NIH HHS/United States
- R01-DK60034/DK/NIDDK NIH HHS/United States
- R01 DK060034/DK/NIDDK NIH HHS/United States
- R01-GM097000/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical