Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing - PubMed (original) (raw)
. 2012 Sep 14;150(6):1107-20.
doi: 10.1016/j.cell.2012.08.029.
Alice H Berger, Peter S Hammerman, Bryan Hernandez, Trevor J Pugh, Eran Hodis, Jeonghee Cho, James Suh, Marzia Capelletti, Andrey Sivachenko, Carrie Sougnez, Daniel Auclair, Michael S Lawrence, Petar Stojanov, Kristian Cibulskis, Kyusam Choi, Luc de Waal, Tanaz Sharifnia, Angela Brooks, Heidi Greulich, Shantanu Banerji, Thomas Zander, Danila Seidel, Frauke Leenders, Sascha Ansén, Corinna Ludwig, Walburga Engel-Riedel, Erich Stoelben, Jürgen Wolf, Chandra Goparju, Kristin Thompson, Wendy Winckler, David Kwiatkowski, Bruce E Johnson, Pasi A Jänne, Vincent A Miller, William Pao, William D Travis, Harvey I Pass, Stacey B Gabriel, Eric S Lander, Roman K Thomas, Levi A Garraway, Gad Getz, Matthew Meyerson
Affiliations
- PMID: 22980975
- PMCID: PMC3557932
- DOI: 10.1016/j.cell.2012.08.029
Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing
Marcin Imielinski et al. Cell. 2012.
Abstract
Lung adenocarcinoma, the most common subtype of non-small cell lung cancer, is responsible for more than 500,000 deaths per year worldwide. Here, we report exome and genome sequences of 183 lung adenocarcinoma tumor/normal DNA pairs. These analyses revealed a mean exonic somatic mutation rate of 12.0 events/megabase and identified the majority of genes previously reported as significantly mutated in lung adenocarcinoma. In addition, we identified statistically recurrent somatic mutations in the splicing factor gene U2AF1 and truncating mutations affecting RBM10 and ARID1A. Analysis of nucleotide context-specific mutation signatures grouped the sample set into distinct clusters that correlated with smoking history and alterations of reported lung adenocarcinoma genes. Whole-genome sequence analysis revealed frequent structural rearrangements, including in-frame exonic alterations within EGFR and SIK2 kinases. The candidate genes identified in this study are attractive targets for biological characterization and therapeutic targeting of lung adenocarcinoma.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
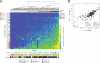
Figure 1. Mutation spectrum analysis of 183 lung adenocarcinomas
(A) Hierarchical clustering of 183 lung adenocarcinomas according to their nucleotide context-specific exonic mutation rates. Each column represents a case, and each row represents one of 96 strand-collapsed trinucleotide context mutation signatures. Top bar: patient-cluster membership. Left bar: simplified single-nucleotide context mutational signature. Bottom bars: reported tumor stage, age, and smoking status for each patient. Right gradient: mutation rate scale. (B) Stratification of reported versus imputed smoking status by the log transform of the adjusted ratio of C->A tranversion rates and CpG->T transition rates. The color of each inner solid point represents the reported smoking status for that particular patient. The color of each outer circle indicates that patient's imputed smoking status as predicted by the classifier. Additional analytic details are provided in the Extended Experimental Procedures.
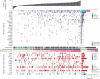
Figure 2. Somatic mutations and copy number changes in 183 lung adenocarcinomas
Top panel, summary of exonic somatic mutation of 25 significantly mutated genes (see text and Table S3 for details). Tumors are arranged from left to right by the number of non-silent mutations per sample, shown in the top track. Significantly mutated genes are listed vertically in decreasing order of non-silent mutation prevalence in the sequenced cohort. Colored rectangles: mutation category observed in a given gene and tumor. Bar chart (right): prevalence of each mutation category in each gene. Asterisks indicate genes significantly enriched in truncating (nonsense, frameshift) mutations. Middle bars: smoking status and mutation spectrum cluster for each patient. White boxes indicate unknown status. Bottom panel: summary of somatic copy number alterations derived from SNP array data. Colored rectangles indicate the copy number change seen for a given gene and tumor.
Figure 3. Somatic mutations of lung adenocarcinoma candidate genes U2AF1, RBM10, and ARID1A
(A) Schematic representation of identified somatic mutations in U2AF1 shown in the context of the known domain structure of the protein. Numbers refer to amino acid residues. Each rectangle corresponds to an independent, mutated tumor sample. Silent mutations are not shown. Missense mutations are shown in black. (B) Schematic of somatic RBM10 mutations. Splice site mutations are shown in purple; truncating mutations are shown in red. Other notations as in (A). (C) Schematic of somatic ARID1A mutations. Notations as in (A) and (B).
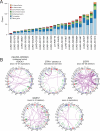
Figure 4. Whole genome sequencing of lung adenocarcinoma
(A) Summary of genic rearrangement types across 25 lung adenocarcinoma whole genomes. Stacked-bar plot depicting the types of somatic rearrangement found in annotated genes by analysis of whole genome sequence data from 25 tumor/normal pairs. The “Other Genic” category refers to rearrangements linking an intergenic region to the 3’ portion of a genic footprint. (B) Representative Circos (Krzywinski et al., 2009) plots of whole genome sequence data with rearrangements targeting known lung adenocarcinoma genes CDKN2A, STK11 and EGFR and novel genes MAST2, SIK2, and ROCK1. Chromosomes are arranged circularly end-to-end with each chromosome's cytobands marked in the outer ring. The inner ring displays copy number data inferred from whole genome sequencing with intrachromosomal events in green and interchromosomal translocations in purple.
Figure 5. Identification of a novel lung adenocarcinoma in-frame deletion in EGFR
(A) Schematic representation of reported EGFR alterations (above protein model) for comparison with a C-terminal deletion event found in this study by whole genome sequencing (below protein model). A schematic depiction of sequencing data shows the expected wild-type reads (gray) in contrast with the observed reads (black) spanning or split by the deletion breakpoint. Supporting paired-end and split read mapping data are shown are Figure S5. (B) Soft agar colony forming assay of NIH-3T3 cells expressing exon 25 and 26-deleted EGFR (Ex25&26del) or wild-type EGFR in the presence or absence of ligand stimulation. The bar graph shows the number of colonies formed by indicated cells with or without EGF in soft agar (n=3, mean +SD). (C) Ex25&26del EGFR is constitutively active in the absence of EGF. The same NIH-3T3 cells used for the assay in (B) were subjected to immunoblotting with anti- phospho-tyrosine (4G10), anti-EGFR and anti-phospho-Akt (S473) antibodies. Blots were probed with anti-Akt and anti-B-actin antibodies (loading control). (D) Cell growth induced by the oncogenic EGFR deletion mutant is suppressed by erlotinib treatment. Ba/F3 cells transformed by either L858R or Ex25&26del mutants were treated with increasing concentrations of erlotinib as indicated for 72 hrs and were assayed for cell viability.
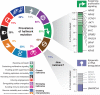
Figure 6. Next-generation hallmarks of lung adenocarcinoma
Left, the prevalence of mutation or SCNA of Sanger Cancer Gene Census (Futreal et al., 2004) genes mapping to cancer hallmarks defined by (Hanahan and Weinberg, 2011). Suspected passenger mutations were filtered out of the analysis, as described in Experimental Procedures. Top right, genes comprising the mutated genes in the hallmark of “sustaining proliferative signaling” are shown. Bottom right, a proposed “11th hallmark” of Epigenetic and RNA deregulation is shown, depicted as above. Genes shown in gray are candidate lung adenocarcinoma genes identified in this study that may additionally contribute to the hallmark.
Similar articles
- Oligogenic germline mutations identified in early non-smokers lung adenocarcinoma patients.
Renieri A, Mencarelli MA, Cetta F, Baldassarri M, Mari F, Furini S, Piu P, Ariani F, Dragani TA, Frullanti E. Renieri A, et al. Lung Cancer. 2014 Aug;85(2):168-74. doi: 10.1016/j.lungcan.2014.05.020. Epub 2014 Jun 4. Lung Cancer. 2014. PMID: 24954872 - Somatic Genomics and Clinical Features of Lung Adenocarcinoma: A Retrospective Study.
Shi J, Hua X, Zhu B, Ravichandran S, Wang M, Nguyen C, Brodie SA, Palleschi A, Alloisio M, Pariscenti G, Jones K, Zhou W, Bouk AJ, Boland J, Hicks B, Risch A, Bennett H, Luke BT, Song L, Duan J, Liu P, Kohno T, Chen Q, Meerzaman D, Marconett C, Laird-Offringa I, Mills I, Caporaso NE, Gail MH, Pesatori AC, Consonni D, Bertazzi PA, Chanock SJ, Landi MT. Shi J, et al. PLoS Med. 2016 Dec 6;13(12):e1002162. doi: 10.1371/journal.pmed.1002162. eCollection 2016 Dec. PLoS Med. 2016. PMID: 27923066 Free PMC article. - Spectrum of gene mutations detected by next generation exome sequencing in brain metastases of lung adenocarcinoma.
Preusser M, Berghoff AS, Koller R, Zielinski CC, Hainfellner JA, Liebmann-Reindl S, Popitsch N, Geier CB, Streubel B, Birner P. Preusser M, et al. Eur J Cancer. 2015 Sep;51(13):1803-11. doi: 10.1016/j.ejca.2015.06.107. Epub 2015 Jul 8. Eur J Cancer. 2015. PMID: 26164066 - Mutation profile of non-small cell lung cancer revealed by next generation sequencing.
Chang YS, Tu SJ, Chen YC, Liu TY, Lee YT, Yen JC, Fang HY, Chang JG. Chang YS, et al. Respir Res. 2021 Jan 6;22(1):3. doi: 10.1186/s12931-020-01608-5. Respir Res. 2021. PMID: 33407425 Free PMC article. - Personalized therapy on the horizon for squamous cell carcinoma of the lung.
Kim HS, Mitsudomi T, Soo RA, Cho BC. Kim HS, et al. Lung Cancer. 2013 Jun;80(3):249-55. doi: 10.1016/j.lungcan.2013.02.015. Epub 2013 Mar 13. Lung Cancer. 2013. PMID: 23489560 Review.
Cited by
- Integrating multi-omics and machine learning survival frameworks to build a prognostic model based on immune function and cell death patterns in a lung adenocarcinoma cohort.
Xie Y, Chen H, Tian M, Wang Z, Wang L, Zhang J, Wang X, Lian C. Xie Y, et al. Front Immunol. 2024 Sep 13;15:1460547. doi: 10.3389/fimmu.2024.1460547. eCollection 2024. Front Immunol. 2024. PMID: 39346927 Free PMC article. - Germline functional variants contribute to somatic mutation and outcomes in neuroblastoma.
Seo ES, Lee JW, Lim J, Shin S, Cho HW, Ju HY, Yoo KH, Sung KW, Park WY. Seo ES, et al. Nat Commun. 2024 Sep 27;15(1):8360. doi: 10.1038/s41467-024-52128-5. Nat Commun. 2024. PMID: 39333105 Free PMC article. - Aberrant pre-mRNA processing in cancer.
Biswas J, Boussi L, Stein E, Abdel-Wahab O. Biswas J, et al. J Exp Med. 2024 Nov 4;221(11):e20230891. doi: 10.1084/jem.20230891. Epub 2024 Sep 24. J Exp Med. 2024. PMID: 39316554 Free PMC article. Review. - U2AF1 S34F enhances tumorigenic potential of lung cells by exhibiting synergy with KRAS mutation and altering response to environmental stress.
Liang CE, Hrabeta-Robinson E, Behera A, Arevalo C, Fetter IJ, Soulette CM, Thornton AM, Sikandar SS, Brooks AN. Liang CE, et al. bioRxiv [Preprint]. 2024 Sep 15:2024.09.11.612492. doi: 10.1101/2024.09.11.612492. bioRxiv. 2024. PMID: 39314447 Free PMC article. Preprint. - Identification of a novel immunogenic death-associated model for predicting the immune microenvironment in lung adenocarcinoma from single-cell and Bulk transcriptomes.
Pan X, Chen H, Zhang L, Xie Y, Zhang K, Lian C, Wang X. Pan X, et al. J Cancer. 2024 Aug 13;15(16):5165-5182. doi: 10.7150/jca.98659. eCollection 2024. J Cancer. 2024. PMID: 39247599 Free PMC article.
References
- Cancer Statistics. W.H. Organization; 2004.
Publication types
MeSH terms
Grants and funding
- U54 HG003067/HG/NHGRI NIH HHS/United States
- CAPMC/ CIHR/Canada
- R01 CA109038/CA/NCI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- T32GM07753/GM/NIGMS NIH HHS/United States
- T32 CA009216/CA/NCI NIH HHS/United States
- T32 CA9216/CA/NCI NIH HHS/United States
- P50 CA090578/CA/NCI NIH HHS/United States
- R01 CA116020/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous