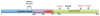Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity - PubMed (original) (raw)
doi: 10.1016/j.eururo.2012.08.053. Epub 2012 Sep 5.
Roman Yelensky, Garrett M Frampton, Kyung Park, Sean R Downing, Theresa Y MacDonald, Mirna Jarosz, Doron Lipson, Scott T Tagawa, David M Nanus, Philip J Stephens, Juan Miguel Mosquera, Maureen T Cronin, Mark A Rubin
Affiliations
- PMID: 22981675
- PMCID: PMC3615043
- DOI: 10.1016/j.eururo.2012.08.053
Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity
Himisha Beltran et al. Eur Urol. 2013 May.
Abstract
Background: Most personalized cancer care strategies involving DNA sequencing are highly reliant on acquiring sufficient fresh or frozen tissue. It has been challenging to comprehensively evaluate the genome of advanced prostate cancer (PCa) because of limited access to metastatic tissue.
Objective: To demonstrate the feasibility of a novel next-generation sequencing (NGS)-based platform that can be used with archival formalin-fixed paraffin-embedded (FFPE) biopsy tissue to evaluate the spectrum of DNA alterations seen in advanced PCa.
Design, setting, and participants: FFPE samples (including archival prostatectomies and prostate needle biopsies) were obtained from 45 patients representing the spectrum of disease: localized PCa, metastatic hormone-naive PCa, and metastatic castration-resistant PCa (CRPC). We also assessed paired primaries and metastases to understand disease heterogeneity and disease progression.
Intervention: At least 50 ng of tumor DNA was extracted from FFPE samples and used for hybridization capture and NGS using the Illumina HiSeq 2000 platform.
Outcome measurements and statistical analysis: A total of 3320 exons of 182 cancer-associated genes and 37 introns of 14 commonly rearranged genes were evaluated for genomic alterations.
Results and limitations: We obtained an average sequencing depth of >900X. Overall, 44% of CRPCs harbored genomic alterations involving the androgen receptor gene (AR), including AR copy number gain (24% of CRPCs) or AR point mutation (20% of CRPCs). Other recurrent mutations included transmembrane protease, serine 2 gene (TMPRSS2):v-ets erythroblastosis virus E26 oncogene homolog (avian) gene (ERG) fusion (44%); phosphatase and tensin homolog gene (PTEN) loss (44%); tumor protein p53 gene (TP53) mutation (40%); retinoblastoma gene (RB) loss (28%); v-myc myelocytomatosis viral oncogene homolog (avian) gene (MYC) gain (12%); and phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α gene (PIK3CA) mutation (4%). There was a high incidence of genomic alterations involving key genes important for DNA repair, including breast cancer 2, early onset gene (BRCA2) loss (12%) and ataxia telangiectasia mutated gene (ATM) mutations (8%); these alterations are potentially targetable with poly(adenosine diphosphate-ribose)polymerase inhibitors. A novel and actionable rearrangement involving the v-raf murine sarcoma viral oncogene homolog B1 gene (BRAF) was also detected.
Conclusions: This first-in-principle study demonstrates the feasibility of performing in-depth DNA analyses using FFPE tissue and brings new insight toward understanding the genomic landscape within advanced PCa.
Copyright © 2012 European Association of Urology. Published by Elsevier B.V. All rights reserved.
Figures
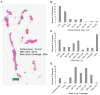
Fig. 1
(a) Representative hematoxylin-eosin photomicrograph of needle core biopsy used for sequencing; (b) tissue surface area for all the samples; (c) DNA yield obtained from samples; (d) mean exon coverage obtained from sequencing.
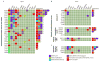
Fig. 2
(a) DNA alterations in castration-resistant prostate cancer (CRPC) (n = 25); (b) DNA alterations in clinically localized prostate cancer (PCa) (n = 16); (c) DNA alterations in metastatic hormone-naive PCa (n = 4). Re-arr = rearrangement; hemiz = hemizygous.
Fig. 3
(a) Clinically localized prostate cancer Gleason grade 6 (3 + 3) with Paneth cell–like neuroendocrine differentiation (upper inset); BRAF-EPB41 gene fusion was identified in this case by sequencing and was validated by fluorescence in situ hybridization(lower inset showing breakapart of the BRAF gene); (b) diagrammatic representation of novel BRAF rearrangement illustrating preserved kinase domain.
Fig. 4
Somatic mutations involving the androgen receptor gene (AR). Mutations marked in black represent novel alterations, whereas mutations marked in red have been previously described as activating point mutations.
Similar articles
- Integrative molecular profiling of routine clinical prostate cancer specimens.
Grasso CS, Cani AK, Hovelson DH, Quist MJ, Douville NJ, Yadati V, Amin AM, Nelson PS, Betz BL, Liu CJ, Knudsen KE, Cooney KA, Feng FY, McDaniel AS, Tomlins SA. Grasso CS, et al. Ann Oncol. 2015 Jun;26(6):1110-1118. doi: 10.1093/annonc/mdv134. Epub 2015 Mar 3. Ann Oncol. 2015. PMID: 25735316 Free PMC article. - Clinical next-generation sequencing in patients with non-small cell lung cancer.
Hagemann IS, Devarakonda S, Lockwood CM, Spencer DH, Guebert K, Bredemeyer AJ, Al-Kateb H, Nguyen TT, Duncavage EJ, Cottrell CE, Kulkarni S, Nagarajan R, Seibert K, Baggstrom M, Waqar SN, Pfeifer JD, Morgensztern D, Govindan R. Hagemann IS, et al. Cancer. 2015 Feb 15;121(4):631-9. doi: 10.1002/cncr.29089. Epub 2014 Oct 24. Cancer. 2015. PMID: 25345567 - Mutation detection in formalin-fixed prostate cancer biopsies taken at the time of diagnosis using next-generation DNA sequencing.
Manson-Bahr D, Ball R, Gundem G, Sethia K, Mills R, Rochester M, Goody V, Anderson E, O'Meara S, Flather M, Keeling M, Yazbek-Hanna M, Hurst R, Curley H, Clark J, Brewer DS, McDermott U, Cooper C. Manson-Bahr D, et al. J Clin Pathol. 2015 Mar;68(3):212-7. doi: 10.1136/jclinpath-2014-202754. Epub 2015 Jan 13. J Clin Pathol. 2015. PMID: 25586381 - Recent advances in prostate cancer research: large-scale genomic analyses reveal novel driver mutations and DNA repair defects.
Frank S, Nelson P, Vasioukhin V. Frank S, et al. F1000Res. 2018 Aug 2;7:F1000 Faculty Rev-1173. doi: 10.12688/f1000research.14499.1. eCollection 2018. F1000Res. 2018. PMID: 30135717 Free PMC article. Review.
Cited by
- Interplay between genomic alterations and androgen receptor signaling during prostate cancer development and progression.
Nyquist MD, Dehm SM. Nyquist MD, et al. Horm Cancer. 2013 Apr;4(2):61-9. doi: 10.1007/s12672-013-0131-4. Epub 2013 Jan 10. Horm Cancer. 2013. PMID: 23307762 Free PMC article. Review. - Luminal breast cancer: from biology to treatment.
Ignatiadis M, Sotiriou C. Ignatiadis M, et al. Nat Rev Clin Oncol. 2013 Sep;10(9):494-506. doi: 10.1038/nrclinonc.2013.124. Epub 2013 Jul 23. Nat Rev Clin Oncol. 2013. PMID: 23881035 Review. - Biallelic BRCA2 Mutations Shape the Somatic Mutational Landscape of Aggressive Prostate Tumors.
Decker B, Karyadi DM, Davis BW, Karlins E, Tillmans LS, Stanford JL, Thibodeau SN, Ostrander EA. Decker B, et al. Am J Hum Genet. 2016 May 5;98(5):818-829. doi: 10.1016/j.ajhg.2016.03.003. Epub 2016 Apr 14. Am J Hum Genet. 2016. PMID: 27087322 Free PMC article. - Identification of key pathways and genes in PTEN mutation prostate cancer by bioinformatics analysis.
Sun J, Li S, Wang F, Fan C, Wang J. Sun J, et al. BMC Med Genet. 2019 Dec 2;20(1):191. doi: 10.1186/s12881-019-0923-7. BMC Med Genet. 2019. PMID: 31791268 Free PMC article. - Preclinical patient-derived modeling of castration-resistant prostate cancer facilitates individualized assessment of homologous recombination repair deficient disease.
Elsesy ME, Oh-Hohenhorst SJ, Oing C, Eckhardt A, Burdak-Rothkamm S, Alawi M, Müller C, Schüller U, Maurer T, von Amsberg G, Petersen C, Rothkamm K, Mansour WY. Elsesy ME, et al. Mol Oncol. 2023 Jun;17(6):1129-1147. doi: 10.1002/1878-0261.13382. Epub 2023 Mar 16. Mol Oncol. 2023. PMID: 36694344 Free PMC article.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. - PubMed
- DeVita VT, Lawrence TS, Rosenberg SA. Cancer: principles and practice of oncology: annual advances in oncology. Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins Health; 2010.
- Beltran H, Beer TM, Carducci MA, et al. New therapies for castration-resistant prostate cancer: efficacy and safety. Eur Urol. 2011;60:279–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous