A maternal high-fat diet modulates fetal SIRT1 histone and protein deacetylase activity in nonhuman primates - PubMed (original) (raw)
A maternal high-fat diet modulates fetal SIRT1 histone and protein deacetylase activity in nonhuman primates
Melissa A Suter et al. FASEB J. 2012 Dec.
Abstract
In nonhuman primates, we previously demonstrated that a maternal high-fat diet (MHFD) induces fetal nonalcoholic fatty liver disease (NAFLD) and alters the fetal metabolome. These changes are accompanied by altered acetylation of histone H3 (H3K14ac). However, the mechanism behind this alteration in acetylation remains unknown. As SIRT1 is both a lysine deacetylase and a crucial sensor of cellular metabolism, we hypothesized that SIRT1 may be involved in fetal epigenomic alterations. Here we show that in utero exposure to a MHFD, but not maternal obesity per se, increases fetal H3K14ac with concomitant decreased SIRT1 expression and diminished in vitro protein and histone deacetylase activity. MHFD increased H3K14ac and DBC1-SIRT1 complex formation in fetal livers, both of which were abrogated with diet reversal despite persistent maternal obesity. Moreover, MHFD was associated with altered expression of known downstream effectors deregulated in NAFLD and modulated by SIRT1 (e.g., PPARΑ, PPARG, SREBF1, CYP7A1, FASN, and SCD). Finally, ex vivo purified SIRT1 retains deacetylase activity on an H3K14ac peptide substrate with preferential activity toward acetylated histone H3; mutagenesis of the catalytic domain of SIRT1 (H363Y) abrogates H3K14ac deacetylation. Our data implicate SIRT1 as a likely molecular mediator of the fetal epigenome and metabolome under MHFD conditions.
Figures
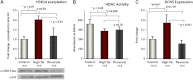
Figure 1.
Fetal hepatic histone H3K14 acetylation, HDAC activity, and HAT expression are altered by virtue of maternal diet exposure. We have previously reported that H3K14 acetylation is increased with exposure to an MHFD. A) Here we show that H3K14ac levels return to those of control-diet levels with diet reversal exposure (_n_=6/group). A representative Western blot is shown at bottom. B) Using a commercially available kit to measure class III HDAC activity, we found that activity is decreased with HFD exposure. Levels remain decreased in diet-reversal animals (_n_=6/group). C) Using qPCR, we measured GCN5 expression levels in fetal hepatic tissue. We found increased GCN5 levels that correspond with increased H3K14 acetylation levels in HFD-exposed animals. GCN5 expression is similar to controls with diet reversal (_n_=10 control, 6 HFD, and 6 reversal). *P < 0.05.
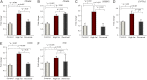
Figure 2.
Fetal hepatic SIRT1 expression, protein, and activity decrease with HFD exposure. A, B) SIRT1 mRNA expression levels (A) and SIRT1 protein levels (B) decrease with HFD exposure. A representative SIRT1 Western blot is included at bottom. C) SIRT1 activity was measured using a commercially available kit (see Materials and Methods) using p53 acetylated substrate (_n_=9 control, 6 HFD, and 7 reversal for A–C). D) DBC1-Sirt1 interaction in HFD-fed primate liver. Equivalent amounts of liver protein homogenates from primates fed either control diet or an HFD or exposed to diet reversal were subjected to immunoprecipitation with Sirt1, followed by Western blot analysis with DBC1 (see Materials and Methods). Fold change relative to control-diet-fed animals expressed relative to IgG input. E) Representative Western blot for DBC1 following anti-SIRT1 immunoprecipitation demonstrates significant increased fetal DBC1 in association with in utero HFD exposure. *P < 0.05.
Figure 3.
SIRT1-associated genes are altered with HFD exposure. A–D) We found that PPARG (A), PPARA (B), SREBF1 (C), and CYP7A1 (D) are all increased with HFD exposure in macaque fetal liver. E) FASN. F) SCD. Except for PPARA (B), whose expression levels remain elevated, all genes show levels similar to control-diet-exposed animals with diet reversal (_n_=10 control, 6 HFD, and 6 reversal). *P < 0.05.
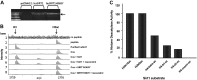
Figure 4.
Catalytically active SIRT1 deacetylates H3K14ac and has preference for acetylated histone H3. A) Cos-1 cells were used for expression and purification of huSIRT1. Cos-1 cells have a low level of expression of endogenous SIRT1, as seen with empty vector (pcDNA3.1), but levels increase with expression of huSIRT1 or huSIRT1H363Y. B) Mass spectrometric analysis identifies H3K14ac as a SIRT1 substrate. Spectrum lanes 1 and 2 shows no-peptide and no-lysate controls, while lane 3 shows that H3K14ac peptide, plus commercially available purified recombinant huSIRT1, demonstrates a shift of −42 Da, indicative of complete deacetylation of K14 by SIRT1. Cos cell lysate (lane 4) without transfected Sirt1 lacks detectable H3K14 deacetylase activity (no −42 Da shift). However, with transfection of hu_Sirt1_ (lane 5) we observe again a −42 Da shift specific to H3K14 deacetylation, and addition of resveratrol (a SIRT1 agonist) to the Cos lysate (lane 6) preserves deacetylation. Transfection with dominant-negative hu_Sirt1_ H363Y into Cos cells without (lane 7) or with (lane 8) resveratrol abrogates this observed deacetylation of H3K14ac. C) Quantitative mass spectrometry shows H3K14ac and H3K9ac as preferential substrates for SIRT1 relative to a tetraacetylated H4 (K5K8K12K16) peptide.
Similar articles
- Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome.
Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, Lane RH. Aagaard-Tillery KM, et al. J Mol Endocrinol. 2008 Aug;41(2):91-102. doi: 10.1677/JME-08-0025. Epub 2008 May 30. J Mol Endocrinol. 2008. PMID: 18515302 Free PMC article. - An acetylation switch modulates the transcriptional activity of estrogen-related receptor alpha.
Wilson BJ, Tremblay AM, Deblois G, Sylvain-Drolet G, Giguère V. Wilson BJ, et al. Mol Endocrinol. 2010 Jul;24(7):1349-58. doi: 10.1210/me.2009-0441. Epub 2010 May 19. Mol Endocrinol. 2010. PMID: 20484414 Free PMC article. - Set7/9 impacts COL2A1 expression through binding and repression of SirT1 histone deacetylation.
Oppenheimer H, Kumar A, Meir H, Schwartz I, Zini A, Haze A, Kandel L, Mattan Y, Liebergall M, Dvir-Ginzberg M. Oppenheimer H, et al. J Bone Miner Res. 2014 Feb;29(2):348-60. doi: 10.1002/jbmr.2052. J Bone Miner Res. 2014. PMID: 23873758 Clinical Trial. - The role of deacetylase SIRT1 in allergic diseases.
Lu Y, Tang X, Wang W, Yang J, Wang S. Lu Y, et al. Front Immunol. 2024 Jul 16;15:1422541. doi: 10.3389/fimmu.2024.1422541. eCollection 2024. Front Immunol. 2024. PMID: 39081309 Free PMC article. Review. - SIRT1 and gynecological malignancies (Review).
Chen J, Chen H, Pan L. Chen J, et al. Oncol Rep. 2021 Apr;45(4):43. doi: 10.3892/or.2021.7994. Epub 2021 Mar 2. Oncol Rep. 2021. PMID: 33649834 Free PMC article. Review.
Cited by
- Developmental Programming of the Fetal Immune System by Maternal Western-Style Diet: Mechanisms and Implications for Disease Pathways in the Offspring.
Nelson BN, Friedman JE. Nelson BN, et al. Int J Mol Sci. 2024 May 29;25(11):5951. doi: 10.3390/ijms25115951. Int J Mol Sci. 2024. PMID: 38892139 Free PMC article. Review. - Initiation of metformin in early pregnancy results in fetal bioaccumulation, growth restriction, and renal dysmorphology in a primate model.
Bolte E, Dean T, Garcia B, Seferovic MD, Sauter K, Hummel G, Bucher M, Li F, Hicks J, Qin X, Suter MA, Barrozo ER, Jochum M, Shope C, Friedman JE, Gannon M, Wesolowski SR, McCurdy CE, Kievit P, Aagaard KM. Bolte E, et al. Am J Obstet Gynecol. 2024 Sep;231(3):352.e1-352.e16. doi: 10.1016/j.ajog.2024.06.002. Epub 2024 Jun 11. Am J Obstet Gynecol. 2024. PMID: 38871238 Free PMC article. - Impact of Transgenerational Nutrition on Nonalcoholic Fatty Liver Disease Development: Interplay between Gut Microbiota, Epigenetics and Immunity.
Tzeng HT, Lee WC. Tzeng HT, et al. Nutrients. 2024 May 3;16(9):1388. doi: 10.3390/nu16091388. Nutrients. 2024. PMID: 38732634 Free PMC article. Review. - Nutrition and Developmental Origins of Kidney Disease.
Nguyen LT, Pollock CA, Saad S. Nguyen LT, et al. Nutrients. 2023 Sep 29;15(19):4207. doi: 10.3390/nu15194207. Nutrients. 2023. PMID: 37836490 Free PMC article. Review. - Maternal cafeteria diet influences kisspeptin (Kiss1), kisspeptin receptor(Gpr54), and sirtuin (Sirt1) genes, hormonal and metabolic profiles, and reproductive functions in rat offspring in a sex-specific manner†.
Matuszewska J, Nowacka-Woszuk J, Radziejewska A, Grzęda E, Pruszyńska-Oszmałek E, Dylewski Ł, Chmurzyńska A, Sliwowska JH. Matuszewska J, et al. Biol Reprod. 2023 Nov 15;109(5):654-668. doi: 10.1093/biolre/ioad101. Biol Reprod. 2023. PMID: 37665248 Free PMC article.
References
- Sauve A. A., Wolberger C., Schramm V. L., Boeke J. D. (2006) The biochemistry of sirtuins. Annu. Rev. Biochem. 75, 435–465 - PubMed
- Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 - PubMed
- Cohen H. Y., Miller C., Bitterman K. J., Wall N. R., Hekking B., Kessler B., Howitz K. T., Gorospe M., de Cabo R., Sinclair D. A. (2004) Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305, 390–392 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K12 GM084897/GM/NIGMS NIH HHS/United States
- R01 DK079194/DK/NIDDK NIH HHS/United States
- R01 DK080558/DK/NIDDK NIH HHS/United States
- DP2 OD001500/OD/NIH HHS/United States
- R01 DA025755/DA/NIDA NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- R01 DK089201/DK/NIDDK NIH HHS/United States
- R24 DK090964/DK/NIDDK NIH HHS/United States
- R01DA025755/DA/NIDA NIH HHS/United States
- DP2120OD001500-01/OD/NIH HHS/United States
- DK79194/DK/NIDDK NIH HHS/United States
- R01DK080558-01/DK/NIDDK NIH HHS/United States
- R01 DK078590/DK/NIDDK NIH HHS/United States
- 5R01DK078590/DK/NIDDK NIH HHS/United States
- R000163/PHS HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous