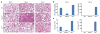A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses - PubMed (original) (raw)
. 2012 Oct 18;490(7420):421-5.
doi: 10.1038/nature11428. Epub 2012 Sep 16.
Affiliations
- PMID: 22982991
- PMCID: PMC3556990
- DOI: 10.1038/nature11428
A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses
Vladimir Litvak et al. Nature. 2012.
Abstract
Antiviral responses must be tightly regulated to defend rapidly against infection while minimizing inflammatory damage. Type 1 interferons (IFN-I) are crucial mediators of antiviral responses and their transcription is regulated by a variety of transcription factors; principal among these is the family of interferon regulatory factors (IRFs). The IRF gene regulatory networks are complex and contain multiple feedback loops. The tools of systems biology are well suited to elucidate the complex interactions that give rise to precise coordination of the interferon response. Here we have used an unbiased systems approach to predict that a member of the forkhead family of transcription factors, FOXO3, is a negative regulator of a subset of antiviral genes. This prediction was validated using macrophages isolated from Foxo3-null mice. Genome-wide location analysis combined with gene deletion studies identified the Irf7 gene as a critical target of FOXO3. FOXO3 was identified as a negative regulator of Irf7 transcription and we have further demonstrated that FOXO3, IRF7 and IFN-I form a coherent feed-forward regulatory circuit. Our data suggest that the FOXO3-IRF7 regulatory circuit represents a novel mechanism for establishing the requisite set points in the interferon pathway that balances the beneficial effects and deleterious sequelae of the antiviral response.
Figures
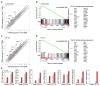
Figure 1. FOXO3 is a negative regulator of the antiviral response
a, Scatter plot comparing global gene expression profiles between unstimulated WT and _Foxo3_-null BMMs. The black lines indicate a two-fold cutoff for the difference in gene expression levels. Data represent the average of three independent experiments. mRNA expression levels are on the log2-scale. b, Gene-set enrichment analysis (GSEA) reveals the overrepresentation of IFN transcriptional signature genes in unstimulated _Foxo3_-null BMMs. Genes are ranked into an ordered list based on relative expression in wild type and _Foxo3_-null BMMs. The middle part of the plot shows the distribution of the genes in the IFN transcriptional signature gene set (“Hits”) against the ranked list of genes. The list on the right shows the top 30 genes in the leading edge subset. Data represent the average of three independent experiments. c, Global gene expression in PIC-stimulated WT and _Foxo3_-null BMMs was analyzed as in a. d, Gene-set enrichment analysis demonstrates up-regulation of IFN transcriptional signature in PIC-stimulated _Foxo3_-null BMMs. Data represent the average of three independent experiments. e, mRNA levels of Cmpk2, Ddx58, Irf7, Isg20, Mx2, Rsad2 and Ifnb1 in WT and _Foxo3_-null macrophages in the presence or absence of PIC stimulation. Data are representative of three experiments (average of three values ± standard error).
Figure 2. FOXO3 keeps the _Irf7_gene in check
a, ChIP-Seq analysis demonstrates FOXO3 binding profile at Irf7 gene promoter in wild type BMMs. Data are representative of two experiments. b, ChIP of FOXO3 from unstimulated wild-type macrophages shows binding of FOXO3 to the promoters of the target genes. FOXO3 recruitment was not observed at control regions lacking FOXO binding sites (-). Data was normalized to IgG (negative control) and represent the average of three independent experiments ± standard error. c, ChIP analysis of histone acetylation, ubiquitination and methylation at Irf7 gene promoter in WT and _Foxo3_-null macrophages. Data represent the average of three independent experiments (± standard error). d, ChIP-Seq analysis demonstrates increased histone H4 acetylation levels in _Foxo3_-null cells. Data are representative of two experiments. e, FOXO3, NCOR2 and HDAC3 are present in the ternary complex at Irf7 promoter, as shown by ChIP-ReChIP assays in unstimulated BMMs. Data was compared to IgG and represent the average of three independent experiments ± standard error. f, ChIP analysis of NCOR2 and HDAC3 binding at Irf7 gene promoter in WT and _Foxo3_-null macrophages. Data represent the average of three independent experiments (± standard error). g, ChIP assay demonstrates increased recruitment of IRF7 at Irf7 gene promoter in _Foxo3_-null macrophages relative. Data was normalized to IgG and represent the average of three independent experiments ± standard error. h, A model depicting the mechanism of FOXO3-mediated repression of Irf7 gene. See text for details.
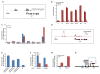
Figure 3. IFNβ represses FOXO3
a, IFNβ- or PIC-stimulation of wild type macrophages was associated with a significant decrease in Foxo3 mRNA levels. PIC-induced decrease of Foxo3 mRNA levels was not observed in _Ifnar1_-null cells. Data are representative of three experiments (average of three values ± standard error). b, IFNβ induces activation of AKT in macrophages. Bar graph demonstrates densitometric quantification of phosphorylated AKT and FOXO3 protein levels. c, IFNβ-induced repression of Foxo3 mRNA levels in WT BMMs was measured in the presence and absence of PI3K and AKT inhibitors. Data are representative of three experiments (average of three values ± standard error). d, A model depicting FOXO3/IRF7/IFN-I regulatory circuit. See text for details.
Figure 4. Antiviral responses lead to increased lung injury in the absence of FOXO3 and IRF7
a, H&E staining of lung tissue sections from wild-type, _Foxo3_-null and _Irf7_-null mice 0, 2 and 5 days after intranasal infection with VSV serotype Indiana 105 p.f.u. Data are from one experiment that is representative of three independent experiments (n=6 mice per group). Scale bar, 200μm. The viral burden in lungs was determined by measurement of VSVg mRNA levels in lung samples using quantitative real-time PCR assay (b) and by standard plaque assays in Vero cells (c). Data are representative of three experiments (average of three values ± standard error).
Similar articles
- MicroRNA-223 Promotes Type I Interferon Production in Antiviral Innate Immunity by Targeting Forkhead Box Protein O3 (FOXO3).
Chen L, Song Y, He L, Wan X, Lai L, Dai F, Liu Y, Wang Q. Chen L, et al. J Biol Chem. 2016 Jul 8;291(28):14706-16. doi: 10.1074/jbc.M115.700252. Epub 2016 May 13. J Biol Chem. 2016. PMID: 27226534 Free PMC article. - Downregulation of miR-155-5p facilitates enterovirus 71 replication through suppression of type I IFN response by targeting FOXO3/IRF7 pathway.
Yang D, Wang X, Gao H, Chen B, Si C, Wang S. Yang D, et al. Cell Cycle. 2020 Jan;19(2):179-192. doi: 10.1080/15384101.2019.1704512. Epub 2019 Dec 19. Cell Cycle. 2020. PMID: 31856677 Free PMC article. - Plasmacytoid dendritic cells control dengue and Chikungunya virus infections via IRF7-regulated interferon responses.
Webster B, Werneke SW, Zafirova B, This S, Coléon S, Décembre E, Paidassi H, Bouvier I, Joubert PE, Duffy D, Walzer T, Albert ML, Dreux M. Webster B, et al. Elife. 2018 Jun 19;7:e34273. doi: 10.7554/eLife.34273. Elife. 2018. PMID: 29914621 Free PMC article. - IRF7: activation, regulation, modification and function.
Ning S, Pagano JS, Barber GN. Ning S, et al. Genes Immun. 2011 Sep;12(6):399-414. doi: 10.1038/gene.2011.21. Epub 2011 Apr 14. Genes Immun. 2011. PMID: 21490621 Free PMC article. Review. - Interferon regulatory factor 7 in inflammation, cancer and infection.
Qing F, Liu Z. Qing F, et al. Front Immunol. 2023 May 12;14:1190841. doi: 10.3389/fimmu.2023.1190841. eCollection 2023. Front Immunol. 2023. PMID: 37251373 Free PMC article. Review.
Cited by
- An Empirical Prior Improves Accuracy for Bayesian Estimation of Transcription Factor Binding Site Frequencies within Gene Promoters.
Ramsey SA. Ramsey SA. Bioinform Biol Insights. 2016 Oct 25;9(Suppl 4):59-69. doi: 10.4137/BBI.S29330. eCollection 2015. Bioinform Biol Insights. 2016. PMID: 27812284 Free PMC article. - Association Between Genetic Variation in FOXO3 and Reductions in Inflammation and Disease Activity in Inflammatory Polyarthritis.
Viatte S, Lee JC, Fu B, Espéli M, Lunt M, De Wolf JN, Wheeler L, Reynolds JA, Castelino M, Symmons DP, Lyons PA, Barton A, Smith KG. Viatte S, et al. Arthritis Rheumatol. 2016 Nov;68(11):2629-2636. doi: 10.1002/art.39760. Arthritis Rheumatol. 2016. PMID: 27214848 Free PMC article. - The methyltransferase Setdb2 mediates virus-induced susceptibility to bacterial superinfection.
Schliehe C, Flynn EK, Vilagos B, Richson U, Swaminanthan S, Bosnjak B, Bauer L, Kandasamy RK, Griesshammer IM, Kosack L, Schmitz F, Litvak V, Sissons J, Lercher A, Bhattacharya A, Khamina K, Trivett AL, Tessarollo L, Mesteri I, Hladik A, Merkler D, Kubicek S, Knapp S, Epstein MM, Symer DE, Aderem A, Bergthaler A. Schliehe C, et al. Nat Immunol. 2015 Jan;16(1):67-74. doi: 10.1038/ni.3046. Epub 2014 Nov 24. Nat Immunol. 2015. PMID: 25419628 Free PMC article. - FAF1 downregulation by Toxoplasma gondii enables host IRF3 mobilization and promotes parasite growth.
Gao FF, Quan JH, Choi IW, Lee YJ, Jang SG, Yuk JM, Lee YH, Cha GH. Gao FF, et al. J Cell Mol Med. 2021 Oct;25(19):9460-9472. doi: 10.1111/jcmm.16889. Epub 2021 Aug 31. J Cell Mol Med. 2021. PMID: 34464509 Free PMC article. - A systems approach to understanding human rhinovirus and influenza virus infection.
Kim TK, Bheda-Malge A, Lin Y, Sreekrishna K, Adams R, Robinson MK, Bascom CC, Tiesman JP, Isfort RJ, Gelinas R. Kim TK, et al. Virology. 2015 Dec;486:146-57. doi: 10.1016/j.virol.2015.08.014. Epub 2015 Oct 6. Virology. 2015. PMID: 26437235 Free PMC article.
References
- Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. - PubMed
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic Acid Recognition by the Innate Immune System. Annu Rev Immunol. 2011;29:185–214. - PubMed
- Benayoun BA, Caburet S, Veitia RA. Forkhead transcription factors: key players in health and disease. Trends Genet. 2011;27:224–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI032972/AI/NIAID NIH HHS/United States
- R01AI032972/AI/NIAID NIH HHS/United States
- U19 AI100627/AI/NIAID NIH HHS/United States
- U54 RR022220/RR/NCRR NIH HHS/United States
- U54GM103511/GM/NIGMS NIH HHS/United States
- HHSN272200800058C/AI/NIAID NIH HHS/United States
- U54 GM103511/GM/NIGMS NIH HHS/United States
- R01AI025032/AI/NIAID NIH HHS/United States
- R01 AI025032/AI/NIAID NIH HHS/United States
- HHSN272200700038C/AI/NIAID NIH HHS/United States
- HSN272200800058C/PHS HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous