OTX2 represses myogenic and neuronal differentiation in medulloblastoma cells - PubMed (original) (raw)
OTX2 represses myogenic and neuronal differentiation in medulloblastoma cells
Ren-Yuan Bai et al. Cancer Res. 2012.
Abstract
The brain development transcription factor OTX2 is overexpressed and/or genomically amplified in most medulloblastomas, but the mechanistic basis for its contributions in this setting are not understood. In this study, we identified OTX2 as a transcriptional repressor and a gatekeeper of myogenic and neuronal differentiation in medulloblastoma cells. OTX2 binds to the MyoD1 core enhancer through its homeobox domain, and the remarkable repressor activity exhibited by the homeobox domain renders OTX2 transcriptionally repressive. RNA interference-mediated attenuation of OTX2 expression triggered myogenic and neuronal differentiation in vitro and prolonged the survival in an orthotopic medulloblastoma mouse model. Conversely, inducing myogenic conversion of medulloblastoma cells led to the loss of OTX2 expression. In medullomyoblastoma, a medulloblastoma subtype containing muscle elements, myogenic cells share cytogenetic signatures with the primitive tumor cells and OTX2 expression was lost in the differentiated myogenic cells. Thus, OTX2 functions via its homeobox domain as a suppressor of differentiation, and the loss of OTX2 expression is linked to the myogenesis in medullomyoblastoma. Together, our findings illustrate the origin of muscle cells in medullomyoblastomas and the oncogenic mechanism of OTX2 as a repressor of diverse differentiating potential.
©2012 AACR.
Conflict of interest statement
Conflict of interest: The authors declare that they have no affiliations that would constitute a financial conflict of interest relating to the subject matter of this study.
Figures
Figure 1. OTX2 knockdown activates myogenic pathway in medulloblastoma cells and OTX2’s association with the core enhancer of MyoD locus
(A) OTX2 knockdown activated myogenic pathway genes and caspase 9. D425 cells were transfected with OTX2 siRNA (OTX2_562) or with randomized control siRNA. Total RNA was isolated from the cells after 36, 48 or 72 h of transfection. RNA of the control (Con) was made after 48 h of the transfection with control siRNA. Gene expression was analyzed by Illumina Beadarray technology and select sets of genes involved in muscle differentiation were displayed in the graph. MyoD (MyoD1): myogenic determination/myogenic factor 3; MyoG: myogenin/myogenic factor 4; MYH3: myosin heavy polypeptide 3, skeletal muscle; MYL4: myosin light polypeptide 4; p21: CDKN1A; GADD45γ: growth arrest and DNA-damage-inducible gamma; MEF2C: MADS box transcription enhancer factor 2; p57: CDKN1C. (B) Neuronal differentiation markers are activated by OTX2 siRNA in D425 cells. As described in (A), select sets of genes involved in neuronal differentiation were displayed. β-TubIII: β-tubulin III; NEF3: neurofilament 3; DCX: doublecortex (doublecortin) transcript variant 3. (C) D425 cells were transfected with OTX2_562 siRNA or a random sequence control siRNA (Con). Western blot (white background) or RT-PCR (black background) was performed at 48, 72 and 96 h for the indicated targets or GAPDH normalization control. Asterix (*) indicates a non-specific band. MYH: skeletal and cardiac myosin heavy chain isoforms. (D) In a similar experiment as (C), medulloblastoma cell line D283 was transfected with OTX2_562 siRNA or a random sequence control siRNA (con). (E) OTX2 siRNA reduces growth in D425 cells and D283 cells. D425 and D283 cells treated by control siRNA, OTX2 siRNAs (OTX2_562) and incubated for 48, 72 or 96 h. Viable cells were quantified by WST-1 reagent and graphed as percentage of the control siRNA cells. (F) Knockdown of OTX2 by siRNA activates the reporter construct of MyoD core enhancer (CER). D425 cells were first transfected with OTX2_562 siRNA or control siRNA for 36 hours. Subsquently, a luciferase construct (pMyoD-CER) containing the 258 bp MyoD CER and a SV40 minimal promoter was transfected and luciferase activity was determined after 24h. (G) Chromatin immunoprecipitation (ChIP) of OTX2 with MyoD CER. ChIP was performed with D425 cells using control rabbit antibody or OTX2 antibody. Bound DNA fragments were detected by PCR with primers encompassing the indicated regions of MyoD locus. (H) OTX2 homeobox domain mediates the interaction with MyoD CER. OTX2 or OTX2 homeobox triple mutant OTX2-3M (R89G, P133T and P134A) was cloned in fusion with VP16 transactivation domain and co-transfected with vector pGL3-P or pMyoD-CER in D425 cells. Luciferase activity was measured and normalized.
Figure 2. OTX2 functions as a transcriptional repressor of MyoD
(A) OTX2 homeobox domain (HD) mediates transcriptional repression. OTX2, OTX2-3M, OTX2-HD (HD) or OTX2 deleted of HD (OTX2-ΔHD) were cloned in fusion with Gal4 DNA binding domain (BD, aa1-147) and co-transfected in D425 cells along with a luciferase reporter construct containing 4x Gal4 binding sites (Gal4-seq). (B) Gal4-OTX2 represses the trans-activity of Gal4-MyoD in D425 cells. Gal4-MyoD was transfected along with empty vector or with indicated Gal4-OTX2 constructs. On the right panel, anti-Gal4 western blot showed the expression levels of transfected constructs. (C) Fusion protein of MyoD and OTX2-HD (HD) showed repressed transcription activity. OTX2-HD (HD), OTX2-HD-3M (HD-3M) or OTX2-ΔHD were cloned in fusion with Gal4-MyoD and tested for their trans-activities in D425 cells. (D) Fusion of OTX2-HD decreases the myogenic potential of MyoD protein in D283 medulloblastoma cells. 3xFLAG-tagged MyoD, MyoD-OTX2-HD (MyoD-HD), MyoD-OTX2-HD-3M (MyoD-HD-3M) or MyoD-OTX2-ΔHD (MyoD-ΔHD) was transiently transfected in D283 cells for 2 days. Cell lysates were blotted for myogenic marker MYH. (E) A summary of the OTX2 mutants used in the study.
Figure 3. OTX2 represses neuronal markers independently of REST
(A) Trichostatin A (TSA) induces neuronal differentiation but not muscle differentiation. D425 cells were incubated with 300 nM of histone deacetylase inhibitor TSA for 48 h and subjected to western blotting analysis of neuronal and muscle differentiation markers. (B) siRNA knockdown of REST induces neuronal differentiation marker β-tubulin III and synapsin I, but shows no effect on cell growth. D425 cells were transfected with control siRNA or REST siRNA in a time course of 48 and 96 h. Western blots revealed the knockdown of REST and the induction of β-tubulin III and synapsin I. (C) OTX2 knockdown greatly increased the activity of synapsin promoter with and without the REST binding site: Re1 motif. Synapsin I promoter with (pSyn) or without (pSyn-ΔRe1) the 23 bp Re1 motif was cloned in pGL3 luciferase construct. D425 cells treated with control siRNA or OTX2_562 siRNA for 36 h were transfected with pGL3 vector, pSyn or pSyn-ΔRe1 for another 36 h. (D) A control experiment of pSyn and pSyn-ΔRe1 showed the Re1 specificity. Neuro2D cells do not express functional REST and Hela cells do. pSyn-ΔRe1 activity was de-repressed compared to pSyn in Hela cells, while both construct showed similar activities in Neuro2D cells. Anti-REST western blot confirmed the REST statues in these cells. (E) Schematic of a proposed model of OTX2 action in medulloblastoma. The homeobox domain of OTX2 acts directly on the core enhancer region (CER) of MyoD to suppress muscle differentiation. Both OTX2 and REST/HDAC are required for the repression of neuronal differentiation markers.
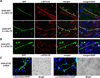
Figure 4. Mutual exclusion of myogenic differentiation and OTX2 expression
(A and B) Loss of OTX2 expression by myogenic conversion of D425 cells. GFP-transfected D425 cells were co-cultured with C2C12 mouse myoblasts in differentiation medium. Cells were stained with myogenic marker MYH or desmin (Des) antibody and Texas Red (TX) secondary antibody (A). Expression of OTX2 was visualized by OTX2 antibody and TX secondary antibody (B). Thirty GFP-positive cells with myotube morphology were examined and none of them showed significant OTX2 staining. White arrow heads indicate the multipile nuclei in the myogenic D245 cells, which were stained by DAPI (merge+DAPI). It is worth noting that the polyclonal D425-GFP cells expressed GFP in various levels, which remained OTX2-positive when maintained in the undifferentiated single cell form. (C) OTX2 knockdown activated myogenic and neuronal differentiation in D425 cells. D425 cells infected with lentivirus of Dox-inducible OTX2-shRNA construct were incubated with 0.1 µg/ml Dox on surface coated with poly-D-lysine for 10 days. Desmin or β-tubulin III staining visualized the cells undergone myogenic or neuronal differentiation. The white arrow head indicates a desmin-positive D425 cell with three nuclei. Bright field pictures with phase contrast were shown next to the immunofluorescent staining. Control D425 cells infected with mock lentivirus did not shown positive staining of desmin and β-tubulin III (data not shown).
Figure 5. OTX2 knockdown induced myogenic marker in D425 xenograft tumor
(A) D425 cells were infected with lentivirus of Dox-inducible OTX2 shRNA. Four days after incubation with Dox, induction of the indicated genes were analyzed by Western blot and RT-PCR. Control cells were infected with the control lentivirus of vector lacking OTX2-shRNA. (B) Survival curve of the nude mice implanted intracranially with D425 cells infected with control lentiviral construct or lentiviral construct with Dox-inducible OTX2 shRNA. Solid lines indicate the groups implanted with control (Con) and OTX2-shRNA cells treated with Dox after 8 days of implantation. Dashed lines indicate the untreated (w/o DOX) mice implanted with control and OTX2-shRNA cells. The mean survival of Con+Dox, OTX2-shRNA+Dox, Con w/o Dox and OTX2-shRNA w/o Dox are 26, 79, 21 and 26.5 days, respectively. (C) Paraffin-embedded brain samples of control D425 xenograft or OTX2 knockdown D425 xenograft were stained by desmin (Des) or OTX2 antibody. 100x objective was used and the areas with colored frames were further magnified and shown on the right side of the picture.
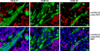
Figure 6. Mutually exclusive expression of OTX2 and myogenic marker desmin in MMBs
Paraffin-embedded MMB samples #1-3 were co-stained with rabbit OTX2 antibody and mouse desmin (Des) antibody, and subsequently with anti-rabbit Texas Red (TX) and anti-mouse FITC secondary antibodies. DAPI staining was merged in the lower panels to indicate the nuclei, where OTX2 is normally located and markedly absent in the desmin-positive cells.
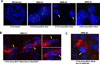
Figure 7. Myogenic cells shares the same cytogenetic signatures with the primitive medulloblastoma tumor cells in MMBs
(A) Hybridization of a normal human leukocyte in metaphase with the OTX2 probe showed two copies of OTX2 locus (left picture). Paraffin-embedded samples of MMB #1–3 were hybridized with _OTX2_-probe labeled in green and c-MYC probed labeled in orange and rendered in red color. DAPI staining revealed the nuclei. (B and C) MMB #1 and #2 samples were first stained with mouse desmin antibody and anti-mouse Cy5 (red) secondary antibody. Subsequently, the sections were hybridized with OTX2 and c-MYC probes and the nuclei were visualized by DAPI (blue). OTX2 signals were rendered in green and c-MYC in white. Yellow arrows indicate the myogenic cells with desmin staining. Enlarged images of the two nuclei indicated by the yellow arrows in B are displayed on the right side of B.
Similar articles
- Characterization of a novel OTX2-driven stem cell program in Group 3 and Group 4 medulloblastoma.
Stromecki M, Tatari N, Morrison LC, Kaur R, Zagozewski J, Palidwor G, Ramaswamy V, Skowron P, Wölfl M, Milde T, Del Bigio MR, Taylor MD, Werbowetski-Ogilvie TE. Stromecki M, et al. Mol Oncol. 2018 Apr;12(4):495-513. doi: 10.1002/1878-0261.12177. Epub 2018 Mar 1. Mol Oncol. 2018. PMID: 29377567 Free PMC article. - OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells.
Bunt J, Hasselt NE, Zwijnenburg DA, Hamdi M, Koster J, Versteeg R, Kool M. Bunt J, et al. Int J Cancer. 2012 Jul 15;131(2):E21-32. doi: 10.1002/ijc.26474. Epub 2011 Nov 8. Int J Cancer. 2012. PMID: 21964830 - Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid.
Di C, Liao S, Adamson DC, Parrett TJ, Broderick DK, Shi Q, Lengauer C, Cummins JM, Velculescu VE, Fults DW, McLendon RE, Bigner DD, Yan H. Di C, et al. Cancer Res. 2005 Feb 1;65(3):919-24. Cancer Res. 2005. PMID: 15705891 - 14q22.3 duplication including OTX2 in a girl with medulloblastoma: A case report with literature review.
Blake C, Widmeyer K, DAquila K, Mochizuki A, Smolarek TA, Pillay-Smiley N, Kim SY. Blake C, et al. Am J Med Genet A. 2024 Jul;194(7):e63604. doi: 10.1002/ajmg.a.63604. Epub 2024 Mar 21. Am J Med Genet A. 2024. PMID: 38511879 Review. - The homeobox gene Otx2 in development and disease.
Beby F, Lamonerie T. Beby F, et al. Exp Eye Res. 2013 Jun;111:9-16. doi: 10.1016/j.exer.2013.03.007. Epub 2013 Mar 21. Exp Eye Res. 2013. PMID: 23523800 Review.
Cited by
- Dissecting super-enhancer driven transcriptional dependencies reveals novel therapeutic strategies and targets for group 3 subtype medulloblastoma.
Li M, Han Y, Wang C, Kang W, Jiang W, Zhang L, Tang Y. Li M, et al. J Exp Clin Cancer Res. 2022 Oct 22;41(1):311. doi: 10.1186/s13046-022-02506-y. J Exp Clin Cancer Res. 2022. PMID: 36273157 Free PMC article. - The landscape of chromatin accessibility in skeletal muscle during embryonic development in pigs.
Yue J, Hou X, Liu X, Wang L, Gao H, Zhao F, Shi L, Shi L, Yan H, Deng T, Gong J, Wang L, Zhang L. Yue J, et al. J Anim Sci Biotechnol. 2021 May 3;12(1):56. doi: 10.1186/s40104-021-00577-z. J Anim Sci Biotechnol. 2021. PMID: 33934724 Free PMC article. - Impact of Reck expression and promoter activity in neuronal in vitro differentiation.
Trombetta-Lima M, Assis-Ribas T, Cintra RC, Campeiro JD, Guerreiro JR, Winnischofer SMB, Nascimento ICC, Ulrich H, Hayashi MAF, Sogayar MC. Trombetta-Lima M, et al. Mol Biol Rep. 2021 Feb;48(2):1985-1994. doi: 10.1007/s11033-021-06175-6. Epub 2021 Feb 22. Mol Biol Rep. 2021. PMID: 33619662 - OTX2 regulates CFTR expression during endoderm differentiation and occupies 3' cis-regulatory elements.
Kerschner JL, Paranjapye A, NandyMazumdar M, Yin S, Leir SH, Harris A. Kerschner JL, et al. Dev Dyn. 2021 May;250(5):684-700. doi: 10.1002/dvdy.293. Epub 2021 Jan 26. Dev Dyn. 2021. PMID: 33386644 Free PMC article. - Otx2 promotes granule cell precursor proliferation and Shh-dependent medulloblastoma maintenance in vivo.
El Nagar S, Chakroun A, Le Greneur C, Figarella-Branger D, Di Meglio T, Lamonerie T, Billon N. El Nagar S, et al. Oncogenesis. 2018 Aug 13;7(8):60. doi: 10.1038/s41389-018-0070-6. Oncogenesis. 2018. PMID: 30100614 Free PMC article.
References
- Klesse LJ, Bowers DC. Childhood medulloblastoma: current status of biology and treatment. CNS Drugs. 24:285–301. - PubMed
- Helton KJ, Fouladi M, Boop FA, Perry A, Dalton J, Kun L, et al. Medullomyoblastoma: a radiographic and clinicopathologic analysis of six cases and review of the literature. Cancer. 2004;101:1445–1454. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials