Involvement of calmodulin and calmodulin kinase II in tumor necrosis factor alpha-induced survival of bone marrow derived macrophages - PubMed (original) (raw)
Involvement of calmodulin and calmodulin kinase II in tumor necrosis factor alpha-induced survival of bone marrow derived macrophages
Jean-Yves Tano et al. Biochem Biophys Res Commun. 2012.
Abstract
We previously showed that survival signaling in TNFα-treated, human THP1-derived macrophages (TDMs) has an obligatory requirement for constitutive Ca(2+) influx through a mechanism involving calmodulin/calmodulin kinase II (CAM/CAMKII). We also demonstrated that such requirement also applies to the protective actions of TNFα in murine bone marrow-derived macrophages (BMDMs) and that TRPC3 channels mediate constitutive Ca(2+) influx. Using a pharmacological approach we here examined if in BMDMs, similarly to TDMs, TNFα-induced survival signaling also involves CAM/CAMKII. In BMDMs, TNFα induced rapid activation of the survival pathways NFκB, AKT and p38MAPK. All these routes were activated in a PI3K-dependent fashion. Activation of AKT and NFκB, but not that of p38MAPK, was abrogated by the CAM inhibitor W7, while KN-62, a CAMKII inhibitor, prevented activation of AKT and p38MAPK but not that of NFκB. Inhibition of CAM or CAMKII completely prevented the protective actions of TNFα. Our observations indicate that in BMDMs CAM and CAMKII have differential contributions to the components of TNFα-dependent survival signaling and underscore a complex interplay among canonical survival routes. These findings set a signaling framework to understand how constitutive Ca(2+) influx couples to macrophage survival in BMDMs.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
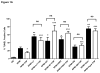
Figure 1
A) Bone marrow-derived macrophages were incubated for 24 hours in complete growth medium (CM), serum-free RPMI medium (RPMI) or RPMI containing TNFα (TNF, 10 ng/ml) in the presence or absence of selective inhibitors of PI3K (LY294002 or LY, 10 μM), CAM (W7, 10 μM), CAMKII (KN62, 25 μM), p38MAPK (SB203580 or SB, 10 μM) or IkBα (hypoestoxide, “Hypo”, 50 μM). Alternatively, cells were incubated (24 h) in RPMI containing those inhibitors but in the absence of TNFα. Following treatments macrophages were processed for evaluation of apoptosis by TUNEL assay (see details in Materials and Methods). *P<0.05, **P<0.0001, respect to RPMI; ***P<0.0003 respect to RPMI+TNF. “ns”: not statistically significant difference. Averages are from four independent experiments. B) Bone marrow-derived macrophages were incubated for 24 hours in serum-free RPMI medium (RPMI) or RPMI containing inhibitors of survival pathways at the concentrations indicated in panel “A”. Following treatments cells were processed for immunodetection of cleaved PARP (89 kDa) in whole cell lysates. Membranes were reprobed for GAPDH to control for protein loading. Shown is a blot representative of three independent experiments and its corresponding densitometric analysis.
Figure 1
A) Bone marrow-derived macrophages were incubated for 24 hours in complete growth medium (CM), serum-free RPMI medium (RPMI) or RPMI containing TNFα (TNF, 10 ng/ml) in the presence or absence of selective inhibitors of PI3K (LY294002 or LY, 10 μM), CAM (W7, 10 μM), CAMKII (KN62, 25 μM), p38MAPK (SB203580 or SB, 10 μM) or IkBα (hypoestoxide, “Hypo”, 50 μM). Alternatively, cells were incubated (24 h) in RPMI containing those inhibitors but in the absence of TNFα. Following treatments macrophages were processed for evaluation of apoptosis by TUNEL assay (see details in Materials and Methods). *P<0.05, **P<0.0001, respect to RPMI; ***P<0.0003 respect to RPMI+TNF. “ns”: not statistically significant difference. Averages are from four independent experiments. B) Bone marrow-derived macrophages were incubated for 24 hours in serum-free RPMI medium (RPMI) or RPMI containing inhibitors of survival pathways at the concentrations indicated in panel “A”. Following treatments cells were processed for immunodetection of cleaved PARP (89 kDa) in whole cell lysates. Membranes were reprobed for GAPDH to control for protein loading. Shown is a blot representative of three independent experiments and its corresponding densitometric analysis.
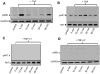
Figure 2
Bone marrow-derived macrophages were treated for the indicated times with TNFα (TNF, 10 ng/ml) in the absence (A, B, E) or presence (C, D, F) of LY294002 (LY, 10 μM) and then processed for immunodetection of phospho-IκBα (Ser32/36, 40 kDa, A, D), phospho-AKT (Ser473; 60 kDa, B, C), or phospho-p38MAPK (pP38, Thr180/Tyr182; 43 kDa, E, F) in whole cell lysates. Membranes were reprobed for GAPDH, total AK or total p38MAPK (P38) to control for protein loading. Blots are representative from three independent experiments.
Figure 2
Bone marrow-derived macrophages were treated for the indicated times with TNFα (TNF, 10 ng/ml) in the absence (A, B, E) or presence (C, D, F) of LY294002 (LY, 10 μM) and then processed for immunodetection of phospho-IκBα (Ser32/36, 40 kDa, A, D), phospho-AKT (Ser473; 60 kDa, B, C), or phospho-p38MAPK (pP38, Thr180/Tyr182; 43 kDa, E, F) in whole cell lysates. Membranes were reprobed for GAPDH, total AK or total p38MAPK (P38) to control for protein loading. Blots are representative from three independent experiments.
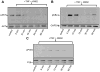
Figure 3
Bone marrow-derived macrophages were pre-incubated for 15 min with the calmodulin inhibitor W7 (25 μM), treated with TNFα (TNF, 10 ng/ml) for the indicated times and then processed for immunodetection of phospho-AKT (Ser473; 60 kDa, A), phospho-IκBα (Ser32/36, 40 kDa, B) or phospho-p38MAPK (pP38, Thr180/Tyr182Ser473; 43 kDa, C) in whole cell lysates. Membranes were reprobed for total AKT, GAPDH or total p38MAPK to control for protein loading. Blots are representative from three independent experiments.
Figure 4
Bone marrow-derived macrophages were pre-incubated for 15 min with the calmodulin-dependent kinase II inhibitor KN62 (10 μM), treated with TNFα (TNF, 10 ng/ml) for the indicated times and then processed for immunodetection of phospho-AKT (Ser473; 60 kDa, A), phospho-IκBα (Ser32/36, 40 kDa, B) or phospho-p38MAPK (pP38, Thr180/Tyr182Ser473; 43 kDa, C) in whole cell lysates. Membranes were reprobed for total AKT, GAPDH or total p38MAPK to control for protein loading. Blots are representative from three independent experiments.
Similar articles
- Requirement for non-regulated, constitutive calcium influx in macrophage survival signaling.
Tano JY, Vazquez G. Tano JY, et al. Biochem Biophys Res Commun. 2011 Apr 8;407(2):432-7. doi: 10.1016/j.bbrc.2011.03.048. Epub 2011 Mar 23. Biochem Biophys Res Commun. 2011. PMID: 21414290 - The KN-93 Molecule Inhibits Calcium/Calmodulin-Dependent Protein Kinase II (CaMKII) Activity by Binding to Ca2+/CaM.
Wong MH, Samal AB, Lee M, Vlach J, Novikov N, Niedziela-Majka A, Feng JY, Koltun DO, Brendza KM, Kwon HJ, Schultz BE, Sakowicz R, Saad JS, Papalia GA. Wong MH, et al. J Mol Biol. 2019 Mar 29;431(7):1440-1459. doi: 10.1016/j.jmb.2019.02.001. Epub 2019 Feb 10. J Mol Biol. 2019. PMID: 30753871 - Impairment of survival signaling and efferocytosis in TRPC3-deficient macrophages.
Tano JY, Smedlund K, Lee R, Abramowitz J, Birnbaumer L, Vazquez G. Tano JY, et al. Biochem Biophys Res Commun. 2011 Jul 8;410(3):643-7. doi: 10.1016/j.bbrc.2011.06.045. Epub 2011 Jun 13. Biochem Biophys Res Commun. 2011. PMID: 21684255 - Calcium, calmodulin, and calcium-calmodulin kinase II: heartbeat to heartbeat and beyond.
Maier LS, Bers DM. Maier LS, et al. J Mol Cell Cardiol. 2002 Aug;34(8):919-39. doi: 10.1006/jmcc.2002.2038. J Mol Cell Cardiol. 2002. PMID: 12234763 Review. - Visualizing CaMKII and CaM activity: a paradigm of compartmentalized signaling.
Bossuyt J, Bers DM. Bossuyt J, et al. J Mol Med (Berl). 2013 Aug;91(8):907-16. doi: 10.1007/s00109-013-1060-y. Epub 2013 Jun 18. J Mol Med (Berl). 2013. PMID: 23775230 Free PMC article. Review.
Cited by
- Pharmacological evidence for a role of the transient receptor potential canonical 3 (TRPC3) channel in endoplasmic reticulum stress-induced apoptosis of human coronary artery endothelial cells.
Ampem PT, Smedlund K, Vazquez G. Ampem PT, et al. Vascul Pharmacol. 2016 Jan;76:42-52. doi: 10.1016/j.vph.2015.07.011. Epub 2015 Jul 26. Vascul Pharmacol. 2016. PMID: 26215710 Free PMC article. - Evidence for a prosurvival role of alpha-7 nicotinic acetylcholine receptor in alternatively (M2)-activated macrophages.
Lee RH, Vazquez G. Lee RH, et al. Physiol Rep. 2013 Dec 26;1(7):e00189. doi: 10.1002/phy2.189. eCollection 2013 Dec 1. Physiol Rep. 2013. PMID: 24744866 Free PMC article. - Reduced endoplasmic reticulum stress-induced apoptosis and impaired unfolded protein response in TRPC3-deficient M1 macrophages.
Solanki S, Dube PR, Tano JY, Birnbaumer L, Vazquez G. Solanki S, et al. Am J Physiol Cell Physiol. 2014 Sep 15;307(6):C521-31. doi: 10.1152/ajpcell.00369.2013. Epub 2014 Jul 16. Am J Physiol Cell Physiol. 2014. PMID: 25031020 Free PMC article. - Targeted inhibition of endothelial calpain delays wound healing by reducing inflammation and angiogenesis.
Yi C, Wu W, Zheng D, Peng G, Huang H, Shen Z, Teng X. Yi C, et al. Cell Death Dis. 2020 Jul 14;11(7):533. doi: 10.1038/s41419-020-02737-x. Cell Death Dis. 2020. PMID: 32665543 Free PMC article. - Propofol attenuates TNF-α-induced MMP-9 expression in human cerebral microvascular endothelial cells by inhibiting Ca2+/CAMK II/ERK/NF-κB signaling pathway.
Ding XW, Sun X, Shen XF, Lu Y, Wang JQ, Sun ZR, Miao CH, Chen JW. Ding XW, et al. Acta Pharmacol Sin. 2019 Oct;40(10):1303-1313. doi: 10.1038/s41401-019-0258-0. Epub 2019 Jun 24. Acta Pharmacol Sin. 2019. PMID: 31235816 Free PMC article.
References
- Gentili C, Picotto G, Morelli S, Boland R, de Boland AR. Effect of ageing in the early biochemical signals elicited by PTH in intestinal cells. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2003;1593:169–178. - PubMed
- Sameermahmood Z, Balasubramanyam M, Saravanan T, Rema M. Curcumin Modulates SDF-1α/CXCR4–Induced Migration of Human Retinal Endothelial Cells (HRECs) Investigative Ophthalmology & Visual Science. 2008;49:3305–3311. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous