Evolution of microbes and viruses: a paradigm shift in evolutionary biology? - PubMed (original) (raw)
Review
Evolution of microbes and viruses: a paradigm shift in evolutionary biology?
Eugene V Koonin et al. Front Cell Infect Microbiol. 2012.
Abstract
When Charles Darwin formulated the central principles of evolutionary biology in the Origin of Species in 1859 and the architects of the Modern Synthesis integrated these principles with population genetics almost a century later, the principal if not the sole objects of evolutionary biology were multicellular eukaryotes, primarily animals and plants. Before the advent of efficient gene sequencing, all attempts to extend evolutionary studies to bacteria have been futile. Sequencing of the rRNA genes in thousands of microbes allowed the construction of the three- domain "ribosomal Tree of Life" that was widely thought to have resolved the evolutionary relationships between the cellular life forms. However, subsequent massive sequencing of numerous, complete microbial genomes revealed novel evolutionary phenomena, the most fundamental of these being: (1) pervasive horizontal gene transfer (HGT), in large part mediated by viruses and plasmids, that shapes the genomes of archaea and bacteria and call for a radical revision (if not abandonment) of the Tree of Life concept, (2) Lamarckian-type inheritance that appears to be critical for antivirus defense and other forms of adaptation in prokaryotes, and (3) evolution of evolvability, i.e., dedicated mechanisms for evolution such as vehicles for HGT and stress-induced mutagenesis systems. In the non-cellular part of the microbial world, phylogenomics and metagenomics of viruses and related selfish genetic elements revealed enormous genetic and molecular diversity and extremely high abundance of viruses that come across as the dominant biological entities on earth. Furthermore, the perennial arms race between viruses and their hosts is one of the defining factors of evolution. Thus, microbial phylogenomics adds new dimensions to the fundamental picture of evolution even as the principle of descent with modification discovered by Darwin and the laws of population genetics remain at the core of evolutionary biology.
Keywords: Darwin; comparative genomics; horizontal gene transfer; modern synthesis; tree of life.
Figures
Figure 1
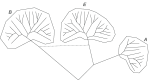
The three-domain tree of life: a generalized schematic, A, Archaea, B, bacteria, E, Eukaryota.
Figure 2
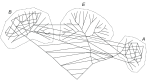
A network representation of the evolutionary process. The network still includes some tree components such that the three domains of cellular life remains distinct but there is also an extensive horizontal component of genetic information flow that in particular dominates the earliest stages of evolution (Koonin and Wolf, 2008).
Figure 3
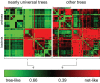
Tree-like (vertical) and web-like (horizontal) contributions in the evolution of nearly universal genes and the entire phylogenetic forest. The two heat maps schematically depict comparison of bacterial and archaeal genomes as described previously (Puigbo et al., 2010).
Figure 4
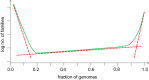
The universal distribution of gene commonality in the microbial genomic universe: a generalized schematic. The three broken lines represent three exponential functions that fit the core (on the right), the shell (in the middle) and the cloud (on the left) of prokaryotic genes (O'Malley and Koonin, 2011).
Figure 5
The core, shell, and cloud of microbial genes. A generalized schematic showing the approximate contributions of the core, shell, and cloud to the pangenomes of prokaryotes and individual genomes.
Figure 6
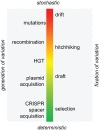
The continuum of evolutionary processes, from stochasticity to determinism.
Figure 7
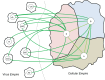
The viral and cellular “empires” of life forms and domains within them. The cellular empire domains: A, Archaea; B, Bacteria; E, Eukaryota. The Virus empire domains: +R, positive-strand RNA viruses; −R, negative-strand RNA viruses; dsR, double-stranded RNA viruses; dsD, double-stranded DNA viruses; ssD, single-stranded DNA viruses; RT, retro-transcribing elements/viruses; VR, viroids.
Figure 8
The conceptual structure of evolutionary biology: the Darwinian core and the new levels of complexity.
Similar articles
- Rhizome of life, catastrophes, sequence exchanges, gene creations, and giant viruses: how microbial genomics challenges Darwin.
Merhej V, Raoult D. Merhej V, et al. Front Cell Infect Microbiol. 2012 Aug 28;2:113. doi: 10.3389/fcimb.2012.00113. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22973559 Free PMC article. Review. - Horizontal gene transfer: essentiality and evolvability in prokaryotes, and roles in evolutionary transitions.
Koonin EV. Koonin EV. F1000Res. 2016 Jul 25;5:F1000 Faculty Rev-1805. doi: 10.12688/f1000research.8737.1. eCollection 2016. F1000Res. 2016. PMID: 27508073 Free PMC article. Review. - Evolution of Microbial Genomics: Conceptual Shifts over a Quarter Century.
Koonin EV, Makarova KS, Wolf YI. Koonin EV, et al. Trends Microbiol. 2021 Jul;29(7):582-592. doi: 10.1016/j.tim.2021.01.005. Epub 2021 Feb 1. Trends Microbiol. 2021. PMID: 33541841 Free PMC article. Review. - Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world.
Koonin EV, Wolf YI. Koonin EV, et al. Nucleic Acids Res. 2008 Dec;36(21):6688-719. doi: 10.1093/nar/gkn668. Epub 2008 Oct 23. Nucleic Acids Res. 2008. PMID: 18948295 Free PMC article. Review. - Evolutionary Genomics of Defense Systems in Archaea and Bacteria.
Koonin EV, Makarova KS, Wolf YI. Koonin EV, et al. Annu Rev Microbiol. 2017 Sep 8;71:233-261. doi: 10.1146/annurev-micro-090816-093830. Epub 2017 Jun 28. Annu Rev Microbiol. 2017. PMID: 28657885 Free PMC article. Review.
Cited by
- Immunity, suicide or both? Ecological determinants for the combined evolution of anti-pathogen defense systems.
Iranzo J, Lobkovsky AE, Wolf YI, Koonin EV. Iranzo J, et al. BMC Evol Biol. 2015 Mar 13;15:43. doi: 10.1186/s12862-015-0324-2. BMC Evol Biol. 2015. PMID: 25881094 Free PMC article. - PADS Arsenal: a database of prokaryotic defense systems related genes.
Zhang Y, Zhang Z, Zhang H, Zhao Y, Zhang Z, Xiao J. Zhang Y, et al. Nucleic Acids Res. 2020 Jan 8;48(D1):D590-D598. doi: 10.1093/nar/gkz916. Nucleic Acids Res. 2020. PMID: 31620779 Free PMC article. - The common ancestor of archaea and eukarya was not an archaeon.
Forterre P. Forterre P. Archaea. 2013;2013:372396. doi: 10.1155/2013/372396. Epub 2013 Nov 17. Archaea. 2013. PMID: 24348094 Free PMC article. Review. - Microbial genomics challenge Darwin.
Raoult D, Koonin EV. Raoult D, et al. Front Cell Infect Microbiol. 2012 Oct 12;2:127. doi: 10.3389/fcimb.2012.00127. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 23091803 Free PMC article. No abstract available. - CLCAs - a family of metalloproteases of intriguing phylogenetic distribution and with cases of substituted catalytic sites.
Lenart A, Dudkiewicz M, Grynberg M, Pawłowski K. Lenart A, et al. PLoS One. 2013 May 9;8(5):e62272. doi: 10.1371/journal.pone.0062272. Print 2013. PLoS One. 2013. PMID: 23671590 Free PMC article.
References
- Arber W. (1979). Promotion and limitation of genetic exchange. Experientia 35, 287–293 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous