IFITM1 is a tight junction protein that inhibits hepatitis C virus entry - PubMed (original) (raw)
IFITM1 is a tight junction protein that inhibits hepatitis C virus entry
Courtney Wilkins et al. Hepatology. 2013 Feb.
Abstract
Type 1 interferon (IFN) continues to be the foundation for the current standard of care combination therapy for chronic hepatitis C virus (HCV) infection, yet the component interferon-stimulated genes (ISGs) that mediate the antiviral actions of IFN are not fully defined. Interferon-induced transmembrane protein 1 (IFITM1) is an ISG product that suppresses early stage infection by a number of viruses through an unknown mechanism of action. Moreover, the actions of IFITM1 on HCV infection are not fully elucidated. Here we identify IFITM1 as a hepatocyte tight junction protein and a potent anti-HCV effector molecule. IFITM1 expression is induced early during IFN treatment of hepatocytes and accumulates at hepatic tight junctions in HCV-infected human patient liver during IFN therapy. Additionally, we found that IFITM1 interacts with HCV coreceptors, including CD81 and occludin, to disrupt the process of viral entry. Thus, IFITM1 is an anti-HCV ISG whose actions impart control of HCV infection through interruption of viral coreceptor function.
Conclusion: This study defines IFITM1 as an ISG effector with action against HCV entry. Design of therapy regimens to enhance IFITM1 expression should improve the virologic response among HCV patients undergoing treatment with type I IFN.
Copyright © 2012 American Association for the Study of Liver Diseases.
Figures
Figure 1
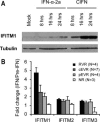
IFITM1 expression associates with effective anti‐HCV responses. (A) Huh7 cells were treated with CIFN or IFN‐α2a for 0, 8, 16, or 24 hours. IFITM1 protein expression was detected via immunoblot assay. (B) RNA was harvested from HCV‐infected patients 24 hours after IFN‐α therapy. Patients were categorized as rapid virologic responders (RVR; undetectable serum HCV RNA by week 4 of IFN therapy), complete or partial early virologic responders (cEVR and pEVR, respectively; undetectable serum HCV RNA or > 2 log10 IU/mL reduction by week 12 of IFN therapy, respectively) or nonresponders (NR; < 2 log10 IU/mL reduction by week 12 of IFN therapy and serum HCV RNA still detectable by week 24) based on level of HCV control by IFN therapy. Quantitative reverse‐transcription polymerase chain reaction (qRT‐PCR) was used to compare IFITM1 levels before and after therapy among these groups.
Figure 2
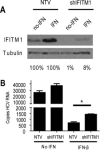
Endogenous IFITM1 contributes to anti‐HCV effects of IFN. (A) Huh7 cells were stably selected for expression of an IFITM1‐specific or nontargeting control shRNA construct. Extracts of cells treated with or without 50 U/mL IFN‐β were probed for IFITM1 expression. Percentages below blots indicate protein levels of IFITM1 relative to nontargeting controls (B) IFITM1 shRNA and nontargeting control Huh7 cells were mock‐treated or treated with 50 U/mL IFN‐β for 16 hours before or 2 hours after HCV infection. Cells were infected with HCV (multiplicity of infection [MOI] = 1), and total RNA harvested at 24 hours postinfection. HCV RNA copy number was quantitated via qRT‐PCR. *P < 0.05.
Figure 3
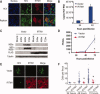
Overexpressed IFITM1 inhibits HCV infection. (A) Huh7 cells or Huh7 cells stably harboring the K2040 HCV subgenomic replicon were transiently transfected with FLAG‐tagged IFITM1 followed by HCV infection (MOI = 1). Viral exclusion was assessed by costaining with HCV patient serum (green) and anti‐FLAG (red) followed by confocal microscopy. Nuclei were visualized via DRAQ5 staining. Images were captured at 60× magnification. (B, C) Huh7 cells stably expressing FLAG‐tagged IFITM1 (Huh7‐IFITM1) or vector control (Huh7‐vector) were infected with HCV (MOI = 0.1) and tested for HCV copy number (via qRT‐PCR) (B) and HCV protein levels (determined via immunoblot assay with anti‐HCV NS5a and anti‐HCV patient serum) (C). (D) Huh7‐IFITM1 and Huh7‐vector were infected with HCV (MOI = 1), and supernatants were collected at the indicated times. Viral titers were assessed via focus‐forming assay on Huh7 cells. (E, F) Huh7‐IFITM1 and Huh7‐vector were infected with HCV (MOI = 0.01). Infected cells were assessed by costaining with HCV patient serum (green) and anti‐FLAG (red) followed by confocal microscopy. (E) Representative focus from each cell type. Images were captured at 60× magnification. (F) Number of HCV‐infected cells per focus. Each point represents one focus, and horizontal lines indicate group means. *P < 0.05.
Figure 4
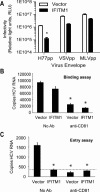
IFITM1 inhibits HCV entry into hepatocytes. (A) Huh7‐IFITM1 and Huh7‐vector were infected with pseudoparticles containing envelopes of the indicated viruses. Viral entry was determined by luciferase activity in relative light units. (B, C) Huh7‐IFITM1 or Huh7‐vector, each with and without neutralizing anti‐CD81, were bound with HCV (MOI = 1) at 4°C for 1 hour. Cells were either harvested for RNA to assess binding (B) or shifted to 37°C for 5 hours, followed by proteinase K cleavage of external virus and subsequent RNA collection to assess entry (C). HCV copy number was determined via qRT‐PCR. The dotted line on the HCV entry panel refers to the limit of detection for this HCV qRT‐PCR assay. *P < 0.05.
Figure 5
IFITM1 alters HCV coreceptor associations. (A) Total cell lysates from Huh7‐IFITM1 or Huh7‐vector were immunoprecipitated as indicated and probed via anti‐FLAG immunoblotting to assess IFITM1 pull‐down. (B) Total cell lysates from Huh7‐IFITM1 and control cells, mock‐ and IFN‐β–treated Huh7 cells, and mock‐ and IFN‐β–treated Huh7 cells harboring nontargeting vector (NTV) or IFITM1‐specific shRNA (shIFITM1) were immunoprecipitated with anti‐CD81 and probed by immunoblot assay assessment of occludin and ZO‐1.
Figure 6
IFITM1 localizes to hepatic tight junctions. (A) Liver tissue from chronically infected HCV patients treated with IFN‐α for 24 hours was costained for IFITM1 (green) and ZO‐1 (red) and visualized using confocal microscopy. (B) Liver tissues from SCID‐uPA chimeric mice treated with IFN‐α for 7 days were costained for IFITM1 in green and tight junctions in red. Samples were visualized using confocal microscopy. (C) Three‐dimensional reconstructions of confocal analysis in panel B. All images were captured at 60× magnification.
Similar articles
- ISG56 and IFITM1 proteins inhibit hepatitis C virus replication.
Raychoudhuri A, Shrivastava S, Steele R, Kim H, Ray R, Ray RB. Raychoudhuri A, et al. J Virol. 2011 Dec;85(24):12881-9. doi: 10.1128/JVI.05633-11. Epub 2011 Oct 5. J Virol. 2011. PMID: 21976647 Free PMC article. - The Interferon-induced Transmembrane Proteins, IFITM1, IFITM2, and IFITM3 Inhibit Hepatitis C Virus Entry.
Narayana SK, Helbig KJ, McCartney EM, Eyre NS, Bull RA, Eltahla A, Lloyd AR, Beard MR. Narayana SK, et al. J Biol Chem. 2015 Oct 23;290(43):25946-59. doi: 10.1074/jbc.M115.657346. Epub 2015 Sep 9. J Biol Chem. 2015. PMID: 26354436 Free PMC article. - Interferon-Induced Transmembrane Protein 1 Restricts Replication of Viruses That Enter Cells via the Plasma Membrane.
Smith SE, Busse DC, Binter S, Weston S, Diaz Soria C, Laksono BM, Clare S, Van Nieuwkoop S, Van den Hoogen BG, Clement M, Marsden M, Humphreys IR, Marsh M, de Swart RL, Wash RS, Tregoning JS, Kellam P. Smith SE, et al. J Virol. 2019 Mar 5;93(6):e02003-18. doi: 10.1128/JVI.02003-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30567988 Free PMC article. - Understanding the molecular mechanism(s) of hepatitis C virus (HCV) induced interferon resistance.
Qashqari H, Al-Mars A, Chaudhary A, Abuzenadah A, Damanhouri G, Alqahtani M, Mahmoud M, El Sayed Zaki M, Fatima K, Qadri I. Qashqari H, et al. Infect Genet Evol. 2013 Oct;19:113-9. doi: 10.1016/j.meegid.2013.06.025. Epub 2013 Jul 5. Infect Genet Evol. 2013. PMID: 23831932 Review. - How hepatitis C virus invades hepatocytes: the mystery of viral entry.
Zhu YZ, Qian XJ, Zhao P, Qi ZT. Zhu YZ, et al. World J Gastroenterol. 2014 Apr 7;20(13):3457-67. doi: 10.3748/wjg.v20.i13.3457. World J Gastroenterol. 2014. PMID: 24707128 Free PMC article. Review.
Cited by
- Antiviral Role of IFITM Proteins in African Swine Fever Virus Infection.
Muñoz-Moreno R, Cuesta-Geijo MÁ, Martínez-Romero C, Barrado-Gil L, Galindo I, García-Sastre A, Alonso C. Muñoz-Moreno R, et al. PLoS One. 2016 Apr 26;11(4):e0154366. doi: 10.1371/journal.pone.0154366. eCollection 2016. PLoS One. 2016. PMID: 27116236 Free PMC article. - IFITMs restrict the replication of multiple pathogenic viruses.
Perreira JM, Chin CR, Feeley EM, Brass AL. Perreira JM, et al. J Mol Biol. 2013 Dec 13;425(24):4937-55. doi: 10.1016/j.jmb.2013.09.024. Epub 2013 Sep 25. J Mol Biol. 2013. PMID: 24076421 Free PMC article. Review. - Regulation of the Interferon Response by lncRNAs in HCV Infection.
Valadkhan S, Fortes P. Valadkhan S, et al. Front Microbiol. 2018 Feb 16;9:181. doi: 10.3389/fmicb.2018.00181. eCollection 2018. Front Microbiol. 2018. PMID: 29503633 Free PMC article. Review. - CD81 and hepatitis C virus (HCV) infection.
Fénéant L, Levy S, Cocquerel L. Fénéant L, et al. Viruses. 2014 Feb 6;6(2):535-72. doi: 10.3390/v6020535. Viruses. 2014. PMID: 24509809 Free PMC article. Review. - Interferon induction of IFITM proteins promotes infection by human coronavirus OC43.
Zhao X, Guo F, Liu F, Cuconati A, Chang J, Block TM, Guo JT. Zhao X, et al. Proc Natl Acad Sci U S A. 2014 May 6;111(18):6756-61. doi: 10.1073/pnas.1320856111. Epub 2014 Apr 21. Proc Natl Acad Sci U S A. 2014. PMID: 24753610 Free PMC article.
References
- Lavanchy D. The global burden of hepatitis C. Liver Int 2009; 29(Suppl. 1): 74‐81. - PubMed
- Kieffer TL, Sarrazin C, Miller JS, Welker MW, Forestier N, Reesink HW, et al. Telaprevir and pegylated interferon‐alpha‐2a inhibit wild‐type and resistant genotype 1 hepatitis C virus replication in patients. HEPATOLOGY 2007; 46: 631‐639. - PubMed
- Gale M Jr. Effector genes of interferon action against hepatitis C virus. HEPATOLOGY 2003; 37: 975‐978. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI060389/AI/NIAID NIH HHS/United States
- DA024563/DA/NIDA NIH HHS/United States
- R01 AI060389/AI/NIAID NIH HHS/United States
- G1100247/MRC_/Medical Research Council/United Kingdom
- U19 AI088778/AI/NIAID NIH HHS/United States
- R01 DK068598/DK/NIDDK NIH HHS/United States
- R01 DA024563/DA/NIDA NIH HHS/United States
- U19 AI040035/AI/NIAID NIH HHS/United States
- G9818340/MRC_/Medical Research Council/United Kingdom
- DK068598-01/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials