Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity - PubMed (original) (raw)
. 2012 Oct;122(10):3692-704.
doi: 10.1172/JCI61623. Epub 2012 Sep 10.
Amy P Hsu, Myung-Jeom Ryu, Jinyong Wang, Xin Gao, Meghan E Boyer, Yangang Liu, Youngsook Lee, Katherine R Calvo, Sunduz Keles, Jing Zhang, Steven M Holland, Emery H Bresnick
Affiliations
- PMID: 22996659
- PMCID: PMC3461907
- DOI: 10.1172/JCI61623
Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity
Kirby D Johnson et al. J Clin Invest. 2012 Oct.
Abstract
Haploinsufficiency for GATA2 causes human immunodeficiency syndromes characterized by mycobacterial infection, myelodysplasia, lymphedema, or aplastic anemia that progress to myeloid leukemia. GATA2 encodes a master regulator of hematopoiesis that is also linked to endothelial biology. Though the disease-causing mutations commonly occur in the GATA-2 DNA binding domain, we identified a patient with mycobacterial infection and myelodysplasia who had an uncharacterized heterozygous deletion in a GATA2 cis-element consisting of an E-box and a GATA motif. Targeted deletion of the equivalent murine element to yield homozygous mutant mice revealed embryonic lethality later than occurred with global Gata2 knockout, hematopoietic stem/progenitor cell depletion, and impaired vascular integrity. Heterozygous mutant mice were viable, but embryos exhibited deficits in definitive, but not primitive, hematopoietic stem/progenitor activity and reduced expression of Gata2 and its target genes. Mechanistic analysis revealed disruption of the endothelial cell transcriptome and loss of vascular integrity. Thus, the composite element disrupted in a human immunodeficiency is essential for establishment of the murine hematopoietic stem/progenitor cell compartment in the fetal liver and for essential vascular processes.
Figures
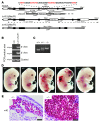
Figure 1. Disruption of the GATA2 E-box–GATA composite _cis_-element in the MonoMAC patient.
(A) ChIP-seq analysis of GATA-2 occupancy at GATA2 in human erythroid, endothelial, and neural cells. Datasets for K562 erythroleukemia and HUVECs were described (27, 43). The ChIP-seq data set from SH-SY5Y neuroblastomas (from R. Kumar, P.J. Farnham, and E.H. Bresnick, unpublished observations) was performed identically to the other analyses. (B) +9.5 kb site deletion in MonoMAC. Sequencing identified a 28-bp deletion from 1 allele of the MonoMAC patient (c.572+512del28 allele) versus control. Human and mouse sequence comparison illustrating conserved composite element. Red text: GATA motifs and E-box (CATCTG). Human transcript variant 1 (NM_001145661.1) includes two upstream noncoding exons. The murine intron 4 +9.5 region corresponds to human intron 5. (C) Co-inheritance of SNP-containing G allele and +9.5 kb site deletion (intron 5 del28) by one son of the MonoMAC patient determined by sequencing, demonstrating linkage of GATA2 +9.5 kb site deletion and exon 4 SNP rs34799090 (c.481C>G). (D) Differential allele expression in the +9.5 kb site deletion MonoMAC patient. SNP probes distinguished between GATA2 alleles in monocyte and T cell cDNAs (mean ± SD, 2 RT reactions per sample analyzed in quadruplicate; *P < 0.0001). (E) Bone marrow megakaryocytic dysplasia from the +9.5 kb site deletion MonoMAC patient versus normal human marrow megakaryocytes. Normal human megakaryocytes from H&E-stained core marrow biopsy and Wright-Giemsa–stained aspirate smear demonstrating normal maturation with multiple connected nuclear lobes. Dysplastic megakaryocytes from +9.5 kb site deletion patient with thrombocytopenia showing atypical separated or detached nuclear lobes (core biopsy, H&E; aspirate smear, Wright-Giemsa) and atypical mononuclear megakaryocyte; dysplastic megakaryocyte detected by Factor VIIIvw immunohistochemistry. Original magnification, ×1000.
Figure 2. Targeted deletion of the +9.5 kb site yields embryonic lethality and severe hemorrhaging.
(A) Strategy for targeted disruption of the composite E-box–GATA element within the +9.5 kb site in murine CJ9 ES cells. GATA motifs and E-box are indicated in red. (B) PCR validation of Δ+9.5 (NeoR+) targeted allele. (C) PCR genotyping assay for Δ+9.5(NeoR)–excised allele. (D) Representative E13.5 littermates. (E) Subcutaneous hemorrhage in the trunk (left) and in the brain ventricular space (right) in +9.5–/– embryos detected by H&E staining of transverse sections. Scale bars: 50 μM.
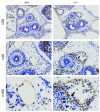
Figure 3. Vascular integrity defect in +9.5–/– E13.5 mouse embryos revealed by PECAM-1 immunohistochemistry.
Representative venous malformation (arrows, ×40 and ×100) and blood cell leakage (arrowheads, ×100) in a transverse section from a +9.5–/– mutant embryo. Blood cells reside both inside and outside the discontinuous endothelial cell layer.
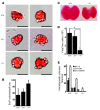
Figure 4. Reduced colony-forming potential of +9.5–/– and +9.5+/– fetal liver but not yolk sac hematopoietic progenitors.
(A) Representative EryP colonies derived from E8.0 yolk sac hematopoietic progenitors cultured in methylcellulose medium for 5 days. Scale bar: 50 μM. (B) Quantitative analysis of EryP colonies obtained from a single litter (+9.5+/+, 3 embryos; +9.5+/–, 3 embryos; +9.5–/–, 3 embryos; mean ± SEM). Differences in the number of colonies from each embryo were not statistically significant. (C) Representative E13.5 fetal livers from +9.5+/+ and +9.5–/– mutant embryos. (D) Quantitative analysis of cell number in E12.5 fetal livers (mean ± SEM; *P < 0.0001). (E) Quantitative analysis of erythroid and myeloid colony-forming potential of fetal liver hematopoietic precursors (3 litters: +9.5+/+, 8 embryos; +9.5+/–, 12 embryos; +9.5–/–, 5 embryos; mean ± SEM). The reduction in erythroid and myeloid colony-forming activity in the +9.5–/– and +9.5+/– samples was significant (P < 0.05). BFU-E, erythroid burst-forming units; GM, granulocyte, monocyte; GEMM, granulocyte, erythrocyte, monocyte, megakaryocyte.
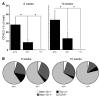
Figure 5. Severe depletion of immunodetectable HSPCs in +9.5–/– embryos.
(A) Flow cytometry analysis of E12.5 fetal livers for HSPC (Lin–Kit+Sca+ and Lin–Kit+) and erythroid cell (TER-119+) content. (B) Cell viability as measured by PI permeability (*P < 0.008). (C) Flow cytometry analysis of E12.5 fetal liver HSCs (Lin–Mac1+CD41–CD48–CD150+Sca+Kit+). (D) Quantitative analysis of flow cytometry data expressed as percentage of total fetal liver cells. *P < 0.005. The analyses conducted in A and B used 4 litters (+9.5+/+, 7 embryos; +9.5+/–, 14 embryos; +9.5–/–, 6 embryos), and analyses in C used 2 litters (+9.5+/+, 5 embryos; +9.5+/–, 5 embryos; +9.5–/–, 6 embryos). Quantitative data are presented as mean ± SEM.
Figure 6. Severe depletion of functional hematopoietic stem cells in +9.5–/– embryos.
E12.5 fetal liver cells (CD45.2) from +9.5+/+, +9.5+/–, and +9.5–/– littermates were transplanted into irradiated CD45.1 recipient mice at a 1:1 ratio with CD45.2+ bone marrow cells. Peripheral blood was analyzed by flow cytometry 6 and 10 weeks after transplantation. (A) CD45.2+ cells in recipient mice expressed as percentage of total live nucleated cells. Data are presented as mean ± SEM. The contribution from +9.5+/– and +9.5–/– donors was significantly reduced compared with +9.5+/+ littermates as indicated (*P < 0.05). (B) The contribution of +9.5+/+ and +9.5+/– donor-derived CD45.2 cells to peripheral blood monocytes (Mac1+Gr-1–), granulocytes (Mac1+Gr-1+), T cells (Thy1.2+), and B cells (CD19+). At 10 weeks, significant increases in CD45.2+ T cells (P = 0.042) and B cells (P = 0.012) were observed in recipients of fetal liver cell transplants from +9.5+/– donors compared with +9.5+/+ littermate controls. Similar results were obtained in an additional transplantation experiment using 150,000 live nucleated fetal liver cells and 300,000 bone marrow cells (data not shown)
Figure 7. Context-dependent mechanism underlying Gata2 transcription in vivo.
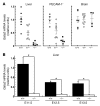
(A) Total RNA was isolated from E12.5–E13.5 liver, endothelial cells, and brain. Gata2 mRNA was quantitated by real-time RT-PCR and normalized by 18S rRNA levels. Each symbol represents the expression level in a particular embryo, and multiple litters: for liver, 2 litters (7 WT, 8 +9.5+/–, 5 mutant); for brain, 3 litters (6 WT, 10 +9.5+/–, 4 mutant) were analyzed. Mean and SEM are represented by long and short horizontal bars, respectively. For all cases except brain, Gata2 expression was significantly lower in mutant versus wild-type samples (+9.5+/+ vs. +9.5–/–, P < 0.0001; +9.5+/+ vs. +9.5+/–, P < 0.04; +9.5+/– vs. +9.5–/–, P < 0.03). (B) Analysis of Gata2 expression in +9.5+/+ vs. +9.5–/– fetal livers from embryos at the indicated developmental stages (+9.5+/+ vs. +9.5–/–, *P < 0.008).
Figure 8. Mechanisms underlying the vascular integrity defect of +9.5–/– embryos.
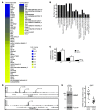
(A) Microarray-based comparison of gene expression in PECAM-1–positive cells purified from E12.5 +9.5+/+ and +9.5–/– embryos. A single-cell suspension was generated from the embryo proper lacking the fetal liver, and PECAM-1–positive cells were isolated by adsorption to magnetic beads loaded with anti–PECAM-1 antibody. Microarray analysis was conducted with four +9.5+/+ and four +9.5–/– embryos, and statistically significant downregulated and upregulated genes are shown in the heat map. (B) Gene ontology analysis of genes downregulated in +9.5–/– embryos using the DAVID Bioinformatics Program (
http://david.abcc.ncifcrf.gov/
). A P value of 0.05 was used as the standard cutoff level. org., organization. (C) Quantitative RT-PCR validation of gene expression changes in PECAM-1+ cells. (D) ChIP-seq profiles of endogenous GATA-2 occupancy of ITGB3 and FERMT3 loci in HUVECs. (E) Itgb3 downregulation in MAE cells upon siRNA-mediated knockdown of GATA-2. Left: Representative Western blot showing the extent of GATA-2 knockdown. Right: Dot plot of Itgb3 mRNA levels in MAE cells treated with control or Gata2 siRNA, respectively. Each dot represents data from a single experiment, and data from 13 independent experiments are shown. Mean and SEM are indicated by long and short horizontal bars, respectively (P < 0.0001).
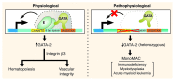
Figure 9. Master regulatory activity of the +9.5 kb _cis_-element to establish the fetal liver HSPC compartment and vascular integrity.
Genetic ablation of the E-box–GATA composite element, which is disrupted as a heterozygous mutation in a MonoMAC patient, severely impaired hematopoiesis and mechanisms that confer vascular integrity. This element is therefore a critical mechanistic determinant of endogenous Gata2 expression in vivo in important target tissues, and its integrity is essential for opposing hematologic disorders.
Similar articles
- Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning.
Lim KC, Hosoya T, Brandt W, Ku CJ, Hosoya-Ohmura S, Camper SA, Yamamoto M, Engel JD. Lim KC, et al. J Clin Invest. 2012 Oct;122(10):3705-17. doi: 10.1172/JCI61619. Epub 2012 Sep 10. J Clin Invest. 2012. PMID: 22996665 Free PMC article. - Human leukemia mutations corrupt but do not abrogate GATA-2 function.
Katsumura KR, Mehta C, Hewitt KJ, Soukup AA, Fraga de Andrade I, Ranheim EA, Johnson KD, Bresnick EH. Katsumura KR, et al. Proc Natl Acad Sci U S A. 2018 Oct 23;115(43):E10109-E10118. doi: 10.1073/pnas.1813015115. Epub 2018 Oct 9. Proc Natl Acad Sci U S A. 2018. PMID: 30301799 Free PMC article. - Functional and molecular characterization of mouse Gata2-independent hematopoietic progenitors.
Kaimakis P, de Pater E, Eich C, Solaimani Kartalaei P, Kauts ML, Vink CS, van der Linden R, Jaegle M, Yokomizo T, Meijer D, Dzierzak E. Kaimakis P, et al. Blood. 2016 Mar 17;127(11):1426-37. doi: 10.1182/blood-2015-10-673749. Epub 2016 Feb 1. Blood. 2016. PMID: 26834239 Free PMC article. - Heterogeneity of GATA2-related myeloid neoplasms.
Hirabayashi S, Wlodarski MW, Kozyra E, Niemeyer CM. Hirabayashi S, et al. Int J Hematol. 2017 Aug;106(2):175-182. doi: 10.1007/s12185-017-2285-2. Epub 2017 Jun 22. Int J Hematol. 2017. PMID: 28643018 Review. - GATA2 +9.5 enhancer: from principles of hematopoiesis to genetic diagnosis in precision medicine.
Soukup AA, Bresnick EH. Soukup AA, et al. Curr Opin Hematol. 2020 May;27(3):163-171. doi: 10.1097/MOH.0000000000000576. Curr Opin Hematol. 2020. PMID: 32205587 Free PMC article. Review.
Cited by
- Oncogenic Enhancers in Leukemia.
Mulet-Lazaro R, Delwel R. Mulet-Lazaro R, et al. Blood Cancer Discov. 2024 Sep 3;5(5):303-317. doi: 10.1158/2643-3230.BCD-23-0211. Blood Cancer Discov. 2024. PMID: 39093124 Review. - PML::RARA and GATA2 proteins interact via DNA templates to induce aberrant self-renewal in mouse and human hematopoietic cells.
Katerndahl CDS, Rogers ORS, Day RB, Xu Z, Helton NM, Ramakrishnan SM, Miller CA, Ley TJ. Katerndahl CDS, et al. Proc Natl Acad Sci U S A. 2024 Apr 30;121(18):e2317690121. doi: 10.1073/pnas.2317690121. Epub 2024 Apr 22. Proc Natl Acad Sci U S A. 2024. PMID: 38648485 Free PMC article. - Functional categorization of gene regulatory variants that cause Mendelian conditions.
Cheng YHH, Bohaczuk SC, Stergachis AB. Cheng YHH, et al. Hum Genet. 2024 Apr;143(4):559-605. doi: 10.1007/s00439-023-02639-w. Epub 2024 Mar 4. Hum Genet. 2024. PMID: 38436667 Free PMC article. Review. - Pathogenic GATA2 genetic variants utilize an obligate enhancer mechanism to distort a multilineage differentiation program.
Katsumura KR, Liu P, Kim JA, Mehta C, Bresnick EH. Katsumura KR, et al. Proc Natl Acad Sci U S A. 2024 Mar 5;121(10):e2317147121. doi: 10.1073/pnas.2317147121. Epub 2024 Feb 29. Proc Natl Acad Sci U S A. 2024. PMID: 38422019 Free PMC article. - Linking GATA2 to myeloid dysplasia and complex cytogenetics in adult myelodysplastic neoplasm and acute myeloid leukemia.
Robbins DJ, Pavletich TS, Patil AT, Pahopos D, Lasarev M, Polaki US, Gahvari ZJ, Bresnick EH, Matson DR. Robbins DJ, et al. Blood Adv. 2024 Jan 9;8(1):80-92. doi: 10.1182/bloodadvances.2023011554. Blood Adv. 2024. PMID: 38029365 Free PMC article.
References
- Tsai F-Y, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89(10):3636–3643. - PubMed
- Zhou Y, Yamamoto M, Engel JD. GATA2 is required for the generation of V2 interneurons. Development. 2000;127(17):3829–3838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK68634/DK/NIDDK NIH HHS/United States
- R01 HG003747/HG/NHGRI NIH HHS/United States
- HG006716/HG/NHGRI NIH HHS/United States
- R21 HG006716/HG/NHGRI NIH HHS/United States
- R01 DK068634/DK/NIDDK NIH HHS/United States
- CA152108/CA/NCI NIH HHS/United States
- R37 DK050107/DK/NIDDK NIH HHS/United States
- R01 DK050107/DK/NIDDK NIH HHS/United States
- P30 CA014520/CA/NCI NIH HHS/United States
- DK50107/DK/NIDDK NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R56 DK068634/DK/NIDDK NIH HHS/United States
- R01 CA152108/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases