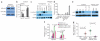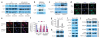LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker - PubMed (original) (raw)
. 2012 Oct;18(10):1511-7.
doi: 10.1038/nm.2940. Epub 2012 Sep 23.
Affiliations
- PMID: 23001183
- PMCID: PMC3684419
- DOI: 10.1038/nm.2940
LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker
Dahu Chen et al. Nat Med. 2012 Oct.
Abstract
There is a pressing need to identify prognostic markers of metastatic disease and targets for treatment. Combining high-throughput RNA sequencing, functional characterization, mechanistic studies and clinical validation, we identify leukemia inhibitory factor receptor (LIFR) as a breast cancer metastasis suppressor downstream of the microRNA miR-9 and upstream of Hippo signaling. Restoring LIFR expression in highly malignant tumor cells suppresses metastasis by triggering a Hippo kinase cascade that leads to phosphorylation, cytoplasmic retention and functional inactivation of the transcriptional coactivator YES-associated protein (YAP). Conversely, loss of LIFR in nonmetastatic breast cancer cells induces migration, invasion and metastatic colonization through activation of YAP. LIFR is downregulated in human breast carcinomas and inversely correlates with metastasis. Notably, in approximately 1,000 nonmetastatic breast tumors, LIFR expression status correlated with metastasis-free, recurrence-free and overall survival outcomes in the patients. These findings identify LIFR as a metastasis suppressor that functions through the Hippo-YAP pathway and has significant prognostic power.
Figures
Figure 1
LIFR is a target of miR-9 and mediates its effect on migration, invasion and metastasis. (a) Immunoblotting of LIFR and β-actin in SUM159 cells infected with the miR-9–expressing vector or empty vector. LE, long exposure; SE, short exposure. (b) Luciferase activity of a reporter fused to a wild-type or mutant LIFR 3′ UTR in SUM159 cells with ectopic expression of miR-9. Mock, mock-infected control cells. (c) Immunoblotting of E-cadherin, LIFR and heat shock protein 90 (HSP90) in human breast cancer cell lines. Metastatic cell lines are defined as cell lines that are capable of launching metastases when growing as primary tumors in mice. (d) Quantitative PCR (qPCR) of miR-9 in the same cell lines used in c. (e) Immunoblotting of LIFR and β-actin in SUM159 cells infected with LIFR shRNA (clones A8 and F3, alone or in combination) and in miR-9–overexpressing SUM159 cells (SUM159_miR-9) infected with human LIFR. Scr, the pGIPZ-GFP lentiviral vector with a scrambled sequence that does not target any mRNA. (f) Transwell migration and Matrigel invasion assays of SUM159 cells infected with LIFR shRNA (shLIFR; clones A8 + F3) and of SUM159_miR-9 cells infected with LIFR. (g) Number of GFP-positive foci in the lungs of mice with orthotopic injection of cells described in f at week 12 after implantation. (n = 10 mice per group). Statistical significance was determined by unpaired, two-tailed Student's t test. Data are means ± s.e.m.
Figure 2
Restoring LIFR expression in highly malignant breast cancer cells suppresses metastasis. (a) H&E staining of primary breast tumors isolated from mice with orthotopic injection of LIFR-transduced 4T1 cells (4T1_LIFR) or mock-infected 4T1 cells (4T1_mock) at day 25 after implantation. Scale bar, 100 μm. The weight of the primary tumor is indicated in parentheses. (b,c) Bright-field imaging (b) and H&E staining (c) of lungs isolated from mice with orthotopic injection of LIFR-transduced or mock-infected 4T1 cells at days 25 and 31 after implantation. Scale bars: b, 2,000 μm; c, 500 μm. Arrows and circles in b indicate visible metastatic nodules. Insets in c are high-magnification (×200) images of specific areas in the corresponding low-magnification (×25) images. (d) Number of metastatic nodules in the lungs of mice with orthotopic injection of LIFR-transduced or mock-infected 4T1 cells at days 25 and 31 after implantation. Data are means ± s.e.m. (n = 7, 9, 10 and 8 mice, respectively, in the four groups shown from left to right). (e,f) Human-specific vimentin immunohistochemical staining (e) and the number of vimentin-positive foci (f) in the lungs of mice with orthotopic injection of LIFR-transduced MDA-MB-231 cells (MDA-MB-231_LIFR) or mock-infected MDA-MB-231 cells (MDA-MB-231_mock) at week 10 after implantation. Scale bar, 600 μm. Insets in e are high-magnification (×600) images of vimentin-positive foci in the corresponding low-magnification (×40) images. Data in f are means ± s.e.m. (n = 9 and 8 mice in the control and LIFR groups, respectively). (g,h) Bright-field imaging (g, left and middle), H&E staining (g, right) and the number of metastatic nodules (h) in the lungs of mice with tail vein injection of LIFR-transduced or mock-infected 4T1 cells at day 21 after implantation. Scale bar, bright-field images (left and middle), 2,000 μm; H&E staining (right), 500 μm. White nodules in g are macroscopic metastases. Data in h are means ± s.e.m. (n = 7 mice per group). Statistical significance was determined by unpaired, two-tailed Student's t test.
Figure 3
LIFR activates Hippo signaling and leads to phosphorylation and functional inactivation of YAP in breast cancer cells. (a) Immunoblotting of phosphorylated YAP (pYAP), YAP and β-actin in mock-infected and LIFR-transduced 4T1 and MDA-MB-231 cells in the presence or absence of LIF stimulation. (b) Immunoblotting of pYAP, YAP and cyclophilin B (CypB) in SUM159 cells infected with LIFR shRNA (shLIFR) or the pGIPZ-GFP lentiviral vector with a scrambled sequence (Scr) that does not target any mRNA. (c) Immunoblotting of YAP, histone H3 (nuclear marker) and HSP90 (cytoplasmic marker) in cytoplasmic and nuclear fractions of 4T1 cells infected with the LIFR-expressing vector or empty vector (mock). The asterisk indicates a nonspecific band that is enriched in the nuclear fraction. (d) Immunofluorescent staining of YAP (green) in mock-infected and LIFR-transduced 4T1 cells. The images on the right are the overlay of YAP and nuclear DAPI (blue) staining of the same field. Scale bar, 20 μm. (e) Immunofluorescent staining of YAP (red) in SUM159 cells infected with the LIFR shRNA (SUM159_shLIFR) or the pGIPZ-GFP lentiviral vector with a scrambled sequence (SUM159_scr). The images on the right are the overlay of YAP and nuclear DAPI (blue) staining of the same field. Scale bar, 20 μm. (f) Percentage of cells with exclusively nuclear (N) YAP and cells with both nuclear and cytoplasmic (N + C) YAP. No cells showed YAP that was localized exclusively in the cytoplasm. Data are means ± s.e.m. (n = 5 fields per group). (g,h) qPCR (g) and immunoblot (h) of CTGF in 4T1 cells infected with the LIFR-expressing vector or empty vector. Data in g are means ± s.e.m. Statistical significance was determined by unpaired, two-tailed Student's t test. (i) Immunoblotting of YAP upstream kinases (phosphorylated MST1 and MST2 (pMST1/2), MST1, phosphorylated LATS1 (pLATS1) and LATS1) in 4T1 cells infected with the LIFR-expressing vector or empty vector. (j) Immunoblotting of total Scribble and plasma membrane-localized Scribble in 4T1 cells infected with the LIFR-expressing vector or empty vector and in SUM159 cells infected with the miR-9–expressing vector or empty vector. Na/K ATPase and HSP90 are markers of the plasma membrane and cytoplasm, respectively. LE, long exposure; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Figure 4
Inhibition of YAP and CTGF mediates the metastasis-suppressing effect of LIFR. (a) Immunoblotting of LIFR, YAP and HSP90 in SUM159 cells transduced with LIFR shRNA (shLIFR) alone or in combination with YAP shRNA (shYAP). (b,c) Bright-field imaging (b) and the number of metastatic nodules (c) in the lungs of mice with tail vein injection of SUM159 cells transduced with LIFR shRNA alone or in combination with YAP shRNA at day 30 after implantation. Scale bar, 2,000 μm. Data in c are means ± s.e.m. (n = 6 mice per group). (d) Immunoblotting of YAP, CTGF and HSP90 in LIFR-expressing 4T1 cells transduced with wild-type (WT) YAP, the S112A YAP mutant or CTGF. (e,f) Primary mammary tumor weight (e) and the number of metastatic nodules in the lungs (f) of mice with orthotopic injection of mock-infected 4T1 cells and LIFR-expressing 4T1 cells transduced with wild-type YAP, the S112A YAP mutant or CTGF at day 22 after implantation. Metastatic nodules were counted under a stereomicroscope and subsequently verified by H&E staining. Data are means ± s.e.m. (n = 9, 10, 10, 9 and 10 mice, respectively, in the five groups shown from left to right). (g) H&E staining of lungs isolated from the mice described in f. Scale bar, 500 μm. Insets are high-magnification images (×200) of specific areas in the corresponding low-magnification (×25) images. Statistical significance was determined by unpaired, two-tailed Student's t test.
Figure 5
LIFR is downregulated in human breast cancer and correlates with clinical outcomes. (a) Immunohistochemical staining of LIFR in representative normal breast, DCIS and invasive breast carcinoma specimens on the NCI Progression TMAs. Brown staining indicates LIFR immunoreactivity. Scale bar, 50 μm. LN, lymph node metastasis. (b) Immunohistochemical staining of LIFR in two representative breast tumor specimens (left, LIFR positive; right, LIFR negative) on the NCI Prognostic TMAs. Brown staining indicates LIFR immunoreactivity. Scale bar, 50 μm. The survival times of both patients are listed. (c–e) Kaplan-Meier graphs representing the probability of cumulative metastasis-free (free of distant metastasis) survival (c), recurrence-free (recurrence indicates tumor relapse at the primary site, the metastatic site or both) survival (d) and overall survival (e) in patients with breast cancer stratified according to LIFR expression status in their primary tumors. Survival time data are presented as means ± s.e.m. (mean survival time is estimated as the area under the survival curve). The log-rank test P value reflects the significance of the correlation between LIFR positivity and longer survival outcome. (f) Model of two metastasis suppressor pathways that are negatively regulated by miR-9 in breast cancer cells. Green indicates oncogenic and/or pro-metastatic factors; pink indicates tumor-suppressing and/or metastasis-suppressing factors. MST1/2, mammalian Hippo homologs 1 and 2; LEF/TCF, lymphoid enhancer-binding factor/T cell-specific factor; LATS1/2, large tumor suppressor homologs 1 and 2; TEAD, TEA domain. “p” in the circles indicates phosphorylation.
Comment in
- LIF-ting Hippo averts metastasis.
Piccolo S. Piccolo S. Nat Med. 2012 Oct;18(10):1463-5. doi: 10.1038/nm.2955. Nat Med. 2012. PMID: 23042347 No abstract available.
Similar articles
- Leukemia inhibitory factor promotes gastric cancer cell proliferation, migration, and invasion via the LIFR-Hippo-YAP pathway.
Bian SB, Yang Y, Liang WQ, Zhang KC, Chen L, Zhang ZT. Bian SB, et al. Ann N Y Acad Sci. 2021 Jan;1484(1):74-89. doi: 10.1111/nyas.14466. Epub 2020 Aug 22. Ann N Y Acad Sci. 2021. PMID: 32827446 - Leukemia Inhibitory Factor Receptor Suppresses the Metastasis of Clear Cell Renal Cell Carcinoma Through Negative Regulation of the Yes-Associated Protein.
Lei C, Lv S, Wang H, Liu C, Zhai Q, Wang S, Cai G, Lu D, Sun Z, Wei Q. Lei C, et al. DNA Cell Biol. 2018 Aug;37(8):659-669. doi: 10.1089/dna.2017.4102. Epub 2018 Jun 14. DNA Cell Biol. 2018. PMID: 29902078 - Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility.
Wang Z, Wu Y, Wang H, Zhang Y, Mei L, Fang X, Zhang X, Zhang F, Chen H, Liu Y, Jiang Y, Sun S, Zheng Y, Li N, Huang L. Wang Z, et al. Proc Natl Acad Sci U S A. 2014 Jan 7;111(1):E89-98. doi: 10.1073/pnas.1319190110. Epub 2013 Dec 23. Proc Natl Acad Sci U S A. 2014. PMID: 24367099 Free PMC article. - Two faces of Hippo: activate or suppress the Hippo pathway in cancer.
Cao J, Huang W. Cao J, et al. Anticancer Drugs. 2017 Nov;28(10):1079-1085. doi: 10.1097/CAD.0000000000000559. Anticancer Drugs. 2017. PMID: 28926422 Review. - Targeting YAP and Hippo signaling pathway in liver cancer.
Liu AM, Xu MZ, Chen J, Poon RT, Luk JM. Liu AM, et al. Expert Opin Ther Targets. 2010 Aug;14(8):855-68. doi: 10.1517/14728222.2010.499361. Expert Opin Ther Targets. 2010. PMID: 20545481 Review.
Cited by
- Mechanosensitive pannexin-1 channels mediate microvascular metastatic cell survival.
Furlow PW, Zhang S, Soong TD, Halberg N, Goodarzi H, Mangrum C, Wu YG, Elemento O, Tavazoie SF. Furlow PW, et al. Nat Cell Biol. 2015 Jul;17(7):943-52. doi: 10.1038/ncb3194. Epub 2015 Jun 22. Nat Cell Biol. 2015. PMID: 26098574 Free PMC article. - The hippo pathway in heart development, regeneration, and diseases.
Zhou Q, Li L, Zhao B, Guan KL. Zhou Q, et al. Circ Res. 2015 Apr 10;116(8):1431-47. doi: 10.1161/CIRCRESAHA.116.303311. Circ Res. 2015. PMID: 25858067 Free PMC article. Review. - The Hippo pathway and human cancer.
Harvey KF, Zhang X, Thomas DM. Harvey KF, et al. Nat Rev Cancer. 2013 Apr;13(4):246-57. doi: 10.1038/nrc3458. Epub 2013 Mar 7. Nat Rev Cancer. 2013. PMID: 23467301 Review. - mTOR pathway occupies a central role in the emergence of latent cancer cells.
Aleksandrova KV, Vorobev ML, Suvorova II. Aleksandrova KV, et al. Cell Death Dis. 2024 Feb 28;15(2):176. doi: 10.1038/s41419-024-06547-3. Cell Death Dis. 2024. PMID: 38418814 Free PMC article. Review. - Morbid Obesity in Women Is Associated with an Altered Intestinal Expression of Genes Related to Cancer Risk and Immune, Defensive, and Antimicrobial Response.
Ho-Plágaro A, Rodríguez-Díaz C, Santiago-Fernández C, López-Gómez C, García-Serrano S, Martín-Reyes F, Rodríguez-Pacheco F, Rodríguez-Cañete A, Alcaín-Martínez G, Vázquez-Pedreño L, Valdés S, Garrido-Sánchez L, García-Fuentes E. Ho-Plágaro A, et al. Biomedicines. 2022 Apr 29;10(5):1024. doi: 10.3390/biomedicines10051024. Biomedicines. 2022. PMID: 35625760 Free PMC article.
References
- Lee YT. Breast carcinoma: pattern of metastasis at autopsy. J. Surg. Oncol. 1983;23:175–180. - PubMed
- Weigelt B, Peterse JL, van `t Veer LJ. Breast cancer metastasis: markers and models. Nat. Rev. Cancer. 2005;5:591–602. - PubMed
- Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat. Med. 2006;12:895–904. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA109311/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- R00 CA138572/CA/NCI NIH HHS/United States
- R00CA138572/CA/NCI NIH HHS/United States
- R01CA166051/CA/NCI NIH HHS/United States
- R01 CA166051/CA/NCI NIH HHS/United States
- CA016672/CA/NCI NIH HHS/United States
- P01CA099031/CA/NCI NIH HHS/United States
- R01CA109311/CA/NCI NIH HHS/United States
- P01 CA099031/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous