Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags - PubMed (original) (raw)
Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags
Yu Wang et al. Appl Environ Microbiol. 2012 Dec.
Abstract
Sediment, a special realm in aquatic environments, has high microbial diversity. While there are numerous reports about the microbial community in marine sediment, freshwater and intertidal sediment communities have been overlooked. The present study determined millions of Illumina reads for a comparison of bacterial communities in freshwater, intertidal wetland, and marine sediments along Pearl River, China, using a technically consistent approach. Our results show that both taxon richness and evenness were the highest in freshwater sediment, medium in intertidal sediment, and lowest in marine sediment. The high number of sequences allowed the determination of a wide variety of bacterial lineages in all sediments for reliable statistical analyses. Principal component analysis showed that the three types of communities could be well separated from phylum to operational taxonomy unit (OTU) levels, and the OTUs from abundant to rare showed satisfactory resolutions. Statistical analysis (LEfSe) demonstrated that the freshwater sediment was enriched with Acidobacteria, Nitrospira, Verrucomicrobia, Alphaproteobacteria, and Betaproteobacteria. The intertidal sediment had a unique community with diverse primary producers (such as Chloroflexi, Bacillariophyta, Gammaproteobacteria, and Epsilonproteobacteria) as well as saprophytic microbes (such as Actinomycetales, Bacteroidetes, and Firmicutes). The marine sediment had a higher abundance of Gammaproteobacteria and Deltaproteobacteria, which were mainly involved in sulfate reduction in anaerobic conditions. These results are helpful for a systematic understanding of bacterial communities in natural sediment environments.
Figures
Fig 1
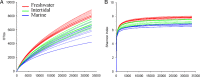
α-Diversity comparison. Rarefaction curves for OTU (A) and Shannon index (B) were calculated using Mothur (v1.27.0) with reads normalized to 33,307 for each sample using 0.03 distance OTUs.
Fig 2
Phylum distribution. (Left) Relative abundance of the dominant bacterial phyla in the three types of sediments. (Right) Principal component analysis (PCA) of phylum abundance data using Canoco 4.5.
Fig 3
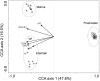
Canonical correspondence analysis (CCA) diagram illustrating the relationship between the genus-level community structure from different sampling sites and environmental variables.
Fig 4
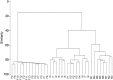
Clustering of samples. Bray-Curis similarity index was calculated using the abundance of OTUs, and hierarchical clustering was calculated using average linkage algorithms using PRIMER 6. F means freshwater samples, I means intertidal samples, and M means marine samples.
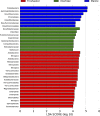
Fig 5
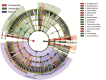
Cladogram indicating the phylogenetic distribution of microbial lineages associated with the three types of sediments; lineages with LDA values of 3.5 or higher determined by LEfSe are displayed. Differences are represented in the color of the most abundant class (red indicating freshwater, green intertidal, purple marine, and yellow nonsignificant). Each circle's diameter is proportional to the taxon's abundance. The strategy of multiclass analysis is nonstrict (at least one class differential). Circles represent phylogenetic levels from domain to genus inside out. Labels are shown of the phylum and class levels.
Fig 6
Indicator microbial groups within the three types of sediments with LDA values higher than 4.
Similar articles
- Comparison among the microbial communities in the lake, lake wetland, and estuary sediments of a plain river network.
Huang W, Chen X, Wang K, Chen J, Zheng B, Jiang X. Huang W, et al. Microbiologyopen. 2019 Feb;8(2):e00644. doi: 10.1002/mbo3.644. Epub 2018 Jun 10. Microbiologyopen. 2019. PMID: 29888529 Free PMC article. - Depth-related changes of sediment ammonia-oxidizing microorganisms in a high-altitude freshwater wetland.
Liu Y, Zhang J, Zhang X, Xie S. Liu Y, et al. Appl Microbiol Biotechnol. 2014 Jun;98(12):5697-707. doi: 10.1007/s00253-014-5651-5. Epub 2014 Mar 12. Appl Microbiol Biotechnol. 2014. PMID: 24619246 - Distinct distribution patterns of prokaryotes between sediment and water in the Yellow River estuary.
Wei G, Li M, Li F, Li H, Gao Z. Wei G, et al. Appl Microbiol Biotechnol. 2016 Nov;100(22):9683-9697. doi: 10.1007/s00253-016-7802-3. Epub 2016 Aug 24. Appl Microbiol Biotechnol. 2016. PMID: 27557722 - Response of microbial community to different media in start-up period of Annan constructed wetland in Beijing of China.
Liu L, Hu J, Teng Y, Wang J, Chen H, Guo X, Zhai Y. Liu L, et al. Environ Pollut. 2023 Nov 15;337:122529. doi: 10.1016/j.envpol.2023.122529. Epub 2023 Sep 8. Environ Pollut. 2023. PMID: 37690468 Review. - Linking Copper-Associated Signal Transduction Systems with Their Environment in Marine Bacteria.
Gautam P, Erill I, Cusick KD. Gautam P, et al. Microorganisms. 2023 Apr 13;11(4):1012. doi: 10.3390/microorganisms11041012. Microorganisms. 2023. PMID: 37110435 Free PMC article. Review.
Cited by
- Soil Fungal Diversity Loss and Appearance of Specific Fungal Pathogenic Communities Associated With the Consecutive Replant Problem (CRP) in Lily.
Shi G, Sun H, Calderón-Urrea A, Jia X, Yang H, Su G. Shi G, et al. Front Microbiol. 2020 Jul 15;11:1649. doi: 10.3389/fmicb.2020.01649. eCollection 2020. Front Microbiol. 2020. PMID: 32760386 Free PMC article. - Archaeal contribution to carbon-functional composition and abundance in China's coastal wetlands: Not to be underestimated.
Yang M, Liu N, Wang B, Li Y, Li J, Liu CQ. Yang M, et al. Front Microbiol. 2022 Nov 10;13:1013408. doi: 10.3389/fmicb.2022.1013408. eCollection 2022. Front Microbiol. 2022. PMID: 36439847 Free PMC article. - Effects of Atractylodes Macrocephala Rhizoma polysaccharide on intestinal microbiota composition in rats with mammary gland hyperplasia.
Ping Y, Li C, Wang L, Zhao H. Ping Y, et al. Front Endocrinol (Lausanne). 2023 Jan 25;13:1102605. doi: 10.3389/fendo.2022.1102605. eCollection 2022. Front Endocrinol (Lausanne). 2023. PMID: 36760814 Free PMC article. - Escherichia coli O101-induced diarrhea develops gut microbial dysbiosis in rats.
Sun X, Gao Y, Wang X, Hu G, Wang Y, Feng B, Hu Y, Mu X, Zhang Y, Dong H. Sun X, et al. Exp Ther Med. 2019 Jan;17(1):824-834. doi: 10.3892/etm.2018.6997. Epub 2018 Nov 20. Exp Ther Med. 2019. PMID: 30651869 Free PMC article. - The effect of plant compartments on the Broussonetia papyrifera-associated fungal and bacterial communities.
Chen P, Zhao M, Tang F, Hu Y, Peng X, Shen S. Chen P, et al. Appl Microbiol Biotechnol. 2020 Apr;104(8):3627-3641. doi: 10.1007/s00253-020-10466-6. Epub 2020 Feb 20. Appl Microbiol Biotechnol. 2020. PMID: 32078018
References
- Antje G, Marc M, Henrik S, Heribert C, Martin K. 2008. Identity and abundance of active sulfate-reducing bacteria in deep tidal flat sediments determined by directed cultivation and CARD-FISH analysis. Environ. Microbiol. 10:2645–2658 - PubMed
- Auguet JC, Barberan A, Casamayor EO. 2010. Global ecological patterns in uncultured archaea. ISME J. 4:182–190 - PubMed
- Barberan A, Casamayor EO. 2010. Global phylogenetic community structure and beta-diversity patterns in surface bacterioplankton metacommunities. Aquat. Microb. Ecol. 59:1–10
- Campbell BJ, Engel AS, Porter ML, Takai K. 2006. The versatile ε-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4:458–468 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources