Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment - PubMed (original) (raw)
doi: 10.1186/gb-2012-13-9-r79.
Timothy L Tickle, Harry Sokol, Dirk Gevers, Kathryn L Devaney, Doyle V Ward, Joshua A Reyes, Samir A Shah, Neal LeLeiko, Scott B Snapper, Athos Bousvaros, Joshua Korzenik, Bruce E Sands, Ramnik J Xavier, Curtis Huttenhower
Affiliations
- PMID: 23013615
- PMCID: PMC3506950
- DOI: 10.1186/gb-2012-13-9-r79
Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment
Xochitl C Morgan et al. Genome Biol. 2012.
Abstract
Background: The inflammatory bowel diseases (IBD) Crohn's disease and ulcerative colitis result from alterations in intestinal microbes and the immune system. However, the precise dysfunctions of microbial metabolism in the gastrointestinal microbiome during IBD remain unclear. We analyzed the microbiota of intestinal biopsies and stool samples from 231 IBD and healthy subjects by 16S gene pyrosequencing and followed up a subset using shotgun metagenomics. Gene and pathway composition were assessed, based on 16S data from phylogenetically-related reference genomes, and associated using sparse multivariate linear modeling with medications, environmental factors, and IBD status.
Results: Firmicutes and Enterobacteriaceae abundances were associated with disease status as expected, but also with treatment and subject characteristics. Microbial function, though, was more consistently perturbed than composition, with 12% of analyzed pathways changed compared with 2% of genera. We identified major shifts in oxidative stress pathways, as well as decreased carbohydrate metabolism and amino acid biosynthesis in favor of nutrient transport and uptake. The microbiome of ileal Crohn's disease was notable for increases in virulence and secretion pathways.
Conclusions: This inferred functional metagenomic information provides the first insights into community-wide microbial processes and pathways that underpin IBD pathogenesis.
Figures
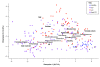
Figure 1
Covariation of microbial community structure in IBD with treatment, environment, biometrics, and disease subtype. Fecal and biopsy samples from 228 IBD patients and healthy controls are plotted as squares (ileal CD) or circles (not ileal involved) and colored by disease status. Axes show the first two components of overall variation as determined by multiple factor analysis (see Materials and methods). Covariation in the presence of clinical factors (bold) and in microbial taxa (italic) is shown. Sample origin (biopsy versus stool) is the single most influential factor in determining microbial community structure, accompanied by host age, treatment types, and disease (particularly ileal CD).
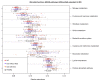
Figure 2
Significant associations of microbial clade abundance and community ecology with IBD and treatment. (a) Taxonomic distribution of clades significant to disease and ileal involvement. Abundant clades not significantly associated with IBD are annotated in gray for context (top 90th percentile of at least 10% of samples and including 5+ genera). Node (non-associated clade) sizes are proportional to the log of the clade's average abundance. (b) Significance of association of sample ecology with disease (CD/UC, ileal/pancolonic), treatment (antibiotics, immunosuppression, mesalamine, steroids), and environment (smoking, stool/biopsy sample origin). Diversity (Simpson's index), evenness (Pielou's index), and richness (Chao1) were calculated for each community (see Materials and methods). False discovery rate q-values are -log10 transformed for visualization, such that values > 0.60 correspond to q < 0.25. Antibiotic treatment is strongly associated with reduced diversity, and stool samples with increased diversity relative to biopsies.
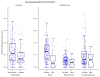
Figure 3
Select microbial clades significantly linked to host environment and treatment. Anaerostipes decreased significantly in the gut communities of smokers, and Dorea, Butyricicoccus, and Coriobacteriaceae were among the taxa most reduced in patients receiving antibiotics (Abx). These associations were significant even in a multivariate model accounting for sample biogeography and disease status. Sqrt, square root.
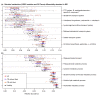
Figure 4
Microbial metabolic pathways with significantly altered abundances in the gut communities of IBD patients. Abundance of KEGG metabolic pathways in microbiome samples is colored by disease state and, when significant, stratified by ileal involvement. Basic metabolism (for example, most amino acid biosynthesis) and SCFA production were reduced in abundance in disease, while biosynthesis and transport of compounds advantageous for oxidative stress (for example, sulfur, cysteine, riboflavin) and adherence/pathogenesis (for example, secretion) were increased.
Figure 5
Small metabolic modules and biological processes with significantly altered abundances in the IBD microbiome. (a, b) Small (typically 5 to 20 gene) KEGG modules (a) and independently defined biological processes from the Gene Ontology (b) were assessed for significant association with disease and ileal involvement as in Figure 4. Metabolism related to oxidative stress (for example, glutathione and sulfate transport) and for pathobiont-like auxotrophy (for example, N-acetylgalactosamine and amino acid uptake) is increased, while several basic biosynthetic processes are less abundant.
Figure 6
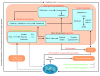
Proposed metabolic roles of the gut microbiome in IBD. Host-mediated processes (blue text) create an environment of oxidative stress in the intestine, which is more favorable to Enterobacteriaceae (increased abundance) than to clades IV and XIVa Clostridia (decreased abundance). This study's inferred IBD metagenomes include broadly increased oxidative metabolism, decreased SCFA production, and increased mucin degradation relative to healthy subjects. These processes all occur within microbes and rely on transport of small molecules to and from the lumen. The resulting tissue-destructive environment provides nutrients such as nucleotides and amino acids, which allow for increased growth of auxotrophic 'specialists'. Bacterial clades of interest are indicated in orange, bacterially mediated processes increased in IBD in red, and processes that decrease in green. Metabolic pathways differential in our IBD communities are contained in blue boxes. GSH and GSSG indicate reduced and oxidized forms of glutathione. LPS, lipopolysaccharide; NAG, N-acetyl galactosamine.
Comment in
- Functional predictions from inference and observation in sequence-based inflammatory bowel disease research.
Meyer F, Trimble WL, Chang EB, Handley KM. Meyer F, et al. Genome Biol. 2012;13(9):169. doi: 10.1186/gb4042. Genome Biol. 2012. PMID: 23013527 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- U54HG004969/HG/NHGRI NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R01 DK092405/DK/NIDDK NIH HHS/United States
- T32 GM080177/GM/NIGMS NIH HHS/United States
- 1R01HG005969/HG/NHGRI NIH HHS/United States
- 1R21HD058828-01A1/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical