Structural evolution of the membrane-coating module of the nuclear pore complex - PubMed (original) (raw)
Structural evolution of the membrane-coating module of the nuclear pore complex
Xiaoping Liu et al. Proc Natl Acad Sci U S A. 2012.
Abstract
The coatomer module of the nuclear pore complex borders the cylinder-like nuclear pore-membrane domain of the nuclear envelope. In evolution, a single coatomer module increases in size from hetero-heptamer (Saccharomyces cerevisiae) to hetero-octamer (Schizosaccharomyces pombe) to hetero-nonamer (Metazoa). Notably, the heptamer-octamer transition proceeds through the acquisition of the nucleoporin Nup37. How Nup37 contacts the heptamer remained unknown. Using recombinant nucleoporins, we show that Sp-Nup37 specifically binds the Sp-Nup120 member of the hetero-heptamer but does not bind an Sc-Nup120 homolog. To elucidate the Nup37-Nup120 interaction at the atomic level, we carried out crystallographic analyses of Sp-Nup37 alone and in a complex with an N-terminal, ~110-kDa fragment of Sp-Nup120 comprising residues 1-950. Corroborating structural predictions, we determined that Nup37 folds into a seven-bladed β-propeller. Several disordered surface regions of the Nup37 β-propeller assume structure when bound to Sp-Nup120. The N-terminal domain of Sp-Nup120(1-950) also folds into a seven-bladed propeller with a markedly protruding 6D-7A insert and is followed by a contorted helical domain. Conspicuously, this 6D-7A insert contains an extension of 50 residues which also is highly conserved in Metazoa but is absent in Sc-Nup120. Strikingly, numerous contacts with the Nup37 β-propeller are located on this extension of the 6D-7A insert. Another contact region is situated toward the end of the helical region of Sp-Nup120(1-950). Our findings provide information about the evolution and the assembly of the coatomer module of the nuclear pore complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
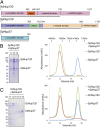
Nup120 of S. pombe (Sp), but not of S. cerevisiae (Sc), dimerizes with Sp-Nup37. (A) Domain structures of Sc-Nup120, Sp-Nup120, and Sp-Nup37 are indicated by various colors; residue numbers indicate boundaries for each domain, and bars depict crystallized domains. (B and C) Full-length Sp-Nup120, but not Sc-Nup120, forms a complex with Sp-Nup37, as detected by gel filtration (Right) and SDS/PAGE analysis of indicated fractions (Left). Asterisk in B indicates major degradation product of Sp-Nup120.
Fig. 2.
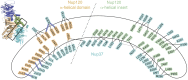
Bicycle-like structure of the Sp-Nup1201–950⋅Sp-Nup37 complex. (A) Cartoon representation of the nonliganded (Upper) and bound (Lower) forms of Nup37 showing the bottom of the β-propeller. Of the four disordered regions (dotted lines, Upper), three assume structure in the bound form (Lower). (B) Ribbon representation from two angles; blue, β-propeller domain of Nup120; green, 6D–7A insert of β-propeller; wheat, helical domain of Nup120; cyan, β-propeller of Nup37. The blades of the two β-propeller structures (note their opposite orientation) and some prominent α-helices are labeled. Note that the helical domain of Nup120 (wheat) provides much of the bicycle frame including one side of each of the two prongs for mounting the two wheels, with the other side provided by the β-propeller insert (green); the “front” wheel is the Nup120 β-propeller; the “rear” wheel is the Nup37 β-propeller. (C) Schematic tracing of polypeptide chains of Sp-Nup1201–950 and full-length Nup37; β-strands of β-propellers (A–E) are indicated by thick arrows; α-helices (h1–h30) are indicated by rectangles and are numbered in order of their occurrence from the N to the C terminus; loops are indicated by lines, and disordered regions are indicated by dotted lines; a dotted outline indicates a portion of β-propeller insert that is absent in Sc-Nup120.
Fig. 4.
Structural comparison of Sp-Nup1201–950 and Sc-Nup1201–729. The two sequences were aligned according to their secondary structures; residue numbers are indicated. Identical residues are shaded in blue; an inserted stretch of about 50 residues in Sp-Nup120 is shaded in pink. Indicated secondary structural elements for Sc-Nup120 are based on the crystal structure determined at 2.6-Å resolution [PDB ID:3F7F (22)]. Color codes are as in Fig. 1_A_. Secondary structural elements are indicated: Arrows indicate β-strands numbered according to their location in the seven-bladed β-propeller; boxes indicate α-helices numbered in order from the N terminus to the C terminus; lines indicate loops, and dotted lines indicate disordered regions. The locations of residues of the Sp-Nup120 interacting with Sp-Nup37 are indicated by green asterisks in the insert domain of the β-propeller and by wheat-colored circles in helical domain.
Fig. 3.
Detailed analyses of the Sp-Nup1201–950⋅Nup37 interactome. Interactions of specific residues by hydrogen bonds (dotted lines), van der Waals interaction (thin lines), and electrostatic interactions (blue lines) are indicated. For reference, a boxed region of the cartoon structure (a miniature equivalent of Fig. 2_A_ , Right) is indicated on the left.
Fig. 5.
Superimposed structures of Sp-Nup1201–950⋅Nup37 and Sc-Nup1201–729. (A) Overlay of the Sp-Nup120 and Sc-Nup120 structures. (B) A 50°-rotated view is shown. β-Propellers (pink for S. cerevisiae and blue for S. pombe) are shown in the Cα skeleton model except for cartoon representations for 1E–1A helix h2 (blue) and helices in the 6D–7A insert (orange for S. cerevisiae and green for S. pombe). Prominent helices are labeled for Sp-Nup120 (lower case) and Sc-Nup120 (upper case). Helical domains are depicted in cartoon model (wheat for S. pombe and pink for S. cerevisiae). Blades of both β-propellers and some helices of inserts and the helical domain are labeled. Nup37 is shown as surface model in light gray. A 21-Å extension of h9 in Sp-Nup120 is indicated by an arrow.
Similar articles
- Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex.
Bilokapic S, Schwartz TU. Bilokapic S, et al. Proc Natl Acad Sci U S A. 2012 Sep 18;109(38):15241-6. doi: 10.1073/pnas.1205151109. Epub 2012 Sep 5. Proc Natl Acad Sci U S A. 2012. PMID: 22955883 Free PMC article. - Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex.
Seo HS, Ma Y, Debler EW, Wacker D, Kutik S, Blobel G, Hoelz A. Seo HS, et al. Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14281-6. doi: 10.1073/pnas.0907453106. Epub 2009 Aug 11. Proc Natl Acad Sci U S A. 2009. PMID: 19706512 Free PMC article. - Nup120: one more piece in the NPC puzzle.
Vetter IR. Vetter IR. Structure. 2009 Aug 12;17(8):1036-8. doi: 10.1016/j.str.2009.07.005. Structure. 2009. PMID: 19679081 - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review. - Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes.
Jaspersen SL, Ghosh S. Jaspersen SL, et al. Nucleus. 2012 May-Jun;3(3):226-36. doi: 10.4161/nucl.20148. Epub 2012 May 1. Nucleus. 2012. PMID: 22572959 Free PMC article. Review.
Cited by
- Integration and deconvolution methodology deciphering prognosis-related signatures in lung adenocarcinoma.
Yi M, Shi J, Tan X, Zhang X, Tao D, Yang Y, Liu Y. Yi M, et al. J Cancer Res Clin Oncol. 2023 Dec;149(18):16441-16460. doi: 10.1007/s00432-023-05403-9. Epub 2023 Sep 14. J Cancer Res Clin Oncol. 2023. PMID: 37710052 - The Structure Inventory of the Nuclear Pore Complex.
Schwartz TU. Schwartz TU. J Mol Biol. 2016 May 22;428(10 Pt A):1986-2000. doi: 10.1016/j.jmb.2016.03.015. Epub 2016 Mar 22. J Mol Biol. 2016. PMID: 27016207 Free PMC article. Review. - Identification of AAAS gene mutation in Allgrove syndrome: A report of three cases.
Li W, Gong C, Qi Z, Wu DI, Cao B. Li W, et al. Exp Ther Med. 2015 Oct;10(4):1277-1282. doi: 10.3892/etm.2015.2677. Epub 2015 Aug 10. Exp Ther Med. 2015. PMID: 26622478 Free PMC article. - Ring cycle for dilating and constricting the nuclear pore.
Solmaz SR, Blobel G, Melcák I. Solmaz SR, et al. Proc Natl Acad Sci U S A. 2013 Apr 9;110(15):5858-63. doi: 10.1073/pnas.1302655110. Epub 2013 Mar 11. Proc Natl Acad Sci U S A. 2013. PMID: 23479651 Free PMC article. - Structure and nucleic acid binding activity of the nucleoporin Nup157.
Seo HS, Blus BJ, Jankovic NZ, Blobel G. Seo HS, et al. Proc Natl Acad Sci U S A. 2013 Oct 8;110(41):16450-5. doi: 10.1073/pnas.1316607110. Epub 2013 Sep 23. Proc Natl Acad Sci U S A. 2013. PMID: 24062435 Free PMC article.
References
- Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–1637. - PubMed
- Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases