c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells - PubMed (original) (raw)
. 2012 Sep 28;151(1):68-79.
doi: 10.1016/j.cell.2012.08.033.
Gangqing Hu, Gang Wei, Kairong Cui, Arito Yamane, Wolfgang Resch, Ruoning Wang, Douglas R Green, Lino Tessarollo, Rafael Casellas, Keji Zhao, David Levens
Affiliations
- PMID: 23021216
- PMCID: PMC3471363
- DOI: 10.1016/j.cell.2012.08.033
c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells
Zuqin Nie et al. Cell. 2012.
Abstract
The c-Myc HLH-bZIP protein has been implicated in physiological or pathological growth, proliferation, apoptosis, metabolism, and differentiation at the cellular, tissue, or organismal levels via regulation of numerous target genes. No principle yet unifies Myc action due partly to an incomplete inventory and functional accounting of Myc's targets. To observe Myc target expression and function in a system where Myc is temporally and physiologically regulated, the transcriptomes and the genome-wide distributions of Myc, RNA polymerase II, and chromatin modifications were compared during lymphocyte activation and in ES cells as well. A remarkably simple rule emerged from this quantitative analysis: Myc is not an on-off specifier of gene activity, but is a nonlinear amplifier of expression, acting universally at active genes, except for immediate early genes that are strongly induced before Myc. This rule of Myc action explains the vast majority of Myc biology observed in literature.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Figure 1. Myc-EGFP expression during lymphocyte activation (see also Figure S1)
(A) Activation and phosphorylation of the c-Myc-EGFP knock-in. Immunoblot analysis of extract from (i) B splenocytes at 0, 1.5, 3.0, 4.5 and 6.0 hours post-LPS activation and (ii) from ConA activated T splenocytes at 0, 1.5, 4, 5, 8, 11, 14 and 18 hours. (B) Flow cytometric analysis of (i) LPS activated c-Myc GFP/GFP splenic B cells at 0, 2, 4, 5, 6 and 7 hours and (ii) of ConA activated c-Myc GFP/GFP splenic T cells at 0, 1.5, 3, 4, 5, 8, 11, 14, 17 and 20 hours. The scale and axes are indicated in the left bottom corner.
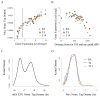
Figure 2. Myc binds to all promoters according to their outputs (see Figure S2 also)
(A) Myc-binding at promoters strongly correlates with expression. Genes were sorted into 20 equal size bins based on expression levels. The averages of Myc ChIP-Seq tag densities at promoters and expression levels are shown for each bin. Dashed vertical lines separate expressed genes from silent or minimally expressed genes. (B) Myc-binding is associated with high expression within ~250 bp of the transcription start site (TSS). Myc targets were sorted into 20 equal-size bins based on the distance of TSS to the nearest peak of c-Myc binding. The averages of gene expression levels (y-axis) and distances (x-axis) are shown for each bin. (C) The distribution of normalized E2F1 ChIP-Seq tag density at mouse ES cell promoters. The y-axis shows the Gaussian kernel density for each tag density point shown in the x-axis. (D) The distribution of normalized Myc tag density at B4, T4 and T14 promoters.
Figure 3. Myc is recruited to promoters according to the amount of RNA polymerase II loaded
(A) Presence (red) and absence (green) of histone markers for heterochromatin (H3K27me2/3, H3K9me2/3 and H4K20me3) and euchromatin (histone acetylation, H2A.Z and H3K4me3) at naive B cell promoters. Genes are sorted by promoter c-Myc tag density. Each line represents a gene. Columns are hierarchically clustered. Only chromosome 1genes are shown. (B) Correlation between normalized c-Myc ChIP-Seq tag density at promoters of B4 cells and RNA Pol II ChIP-Seq tag density at promoters of B resting cells (B0) or B4 cells, with or without 10058-F4 treatment during LPS activation. The promoters are sorted into 20 equal-size groups based on the c-Myc ChIP-Seq tag densities, and the averages of the two sorts of tag densities are shown for each bin. (C) Normalized RNA Pol II ChIP-Seq tag density around TSS in B4 cells treated with or without 10059-F4 during B cell LPS activation. (D) Scatter plot for RNA Pol II pausing index (Muse et al., 2007) in B4 cells treated with and without 10058-F4. (E) The distribution of fold-change of normalized RNA Pol II ChIP-Seq tag density (with 10058-F4/without 10058-F4) in promoter regions (+/− 2K bps around TSS) and in gene body regions (excluding the first 2K bps).
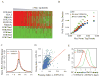
Figure 4. The association of Myc with gene expression, Pol II loading, and open chromatin is conserved in mouse ES cells
(A) Myc binding levels at promoters strongly correlate with gene expression levels (RNA-Seq). Genes were sorted into 20 equal size bins based on gene expression level. Shown for each bin are the averages of Myc ChIP-Seq tag densities (Y-axis) at promoters and of gene expression levels (X-axis). (B) Myc is associated with high gene expression if bound within ~250 bp of TSS. Data analysis as in Figure 2(B). (C) The distribution of normalized Myc tag density at promoters in mouse ES cells. (D) Correlation of Pol II tag densities at promoters and c-Myc ChIP-Seq tag densities at promoters in mouse ES cells. Data analysis as in Figure 3(B). (E) Presence (red) and absence (green) of histone markers for heterochromatin (H3K27me3, H3K9me3 and H4K20me3) and euchromatin (histone acetylation H3K4me) at promoters from mouse ES cells. Genes are sorted by the c-Myc tag density at promoters. Heat-map as in Figure 3(A).
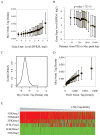
Figure 5. Myc prefers promoters over enhancers
(A) Distribution of Myc ChIP-Seq tag densities (B4 cells) around putative enhancers in resting B cells. Enhancers are defined as p300 binding sites in non-promoter regions. (B) Myc occupancy (B4 cells) at p300 binding sites (resting B cells) is lower in non-promoter vs. promoter regions. (C) Enhancer binding correlates weakly with target expression. The gene nearest an enhancer site is defined as its target. p300 binding sites were sorted by target expression levels. For each group, the average Myc tag density near enhancers (+/− 2K bps) is plotted vs. the average target expression level. The correlation between the levels of Myc enhancer binding and target gene expression was measured by Pearson Coefficient r, in which +1 means a perfect correlation, −1 perfect negative anti-correlation, and 0 no correlation. As a positive control, Myc binding levels at promoters of target genes are plotted against their expression levels for each group. (D), (E), and (F) are similar to (A), (B), (C), respectively, except that the data analysis was done for mouse ES cells.
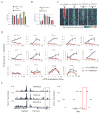
Figure 6. c-Myc amplifies all expressed genes in B-splenocytes (see also Figure S7)
(A) and (B) Flow cytometric analysis of total acridine orange (AO) stained RNA (A) and c-Myc-EGFP (B) in LPS-activated B-splenocytes at 0, 4, 8 and 11 hrs. The cells from wild type (yellow) or c-Myc -EGFP mice were treated with (red), or without (blue) Myc-Max inhibitor 10058-F4. (C) Heatmap of c-Myc tag density (against IgG) near TSS (+/− 2Kbps; 40 windows) for LPS-activated B4 cells treated with and without 10058-F4. Genes are sorted into 100 equal size bins based on expression levels. Shown are the averaged c-Myc tag densities for each bin. The analysis was repeated for p300 binding sites pre-established in resting B cells in non-promoter regions, serving as a proxy for enhancers. The p300 binding sites are sorted into 100 bins based on the H3k27ac level, an estimate of enhancer activity (Creyghton et al., 2010; Rada-Iglesias et al., 2011)). (D) Q-RT-PCR analysis of genes selected randomly from expression array data. Cells were cultured with or without 10058-F4. Two immediate-early genes, Bend3 and Igf2R, expressed with or before c-Myc were 10058-F4 insensitive. Genes Hsd11b1, Bex1 reside in heterochromatin and are inactive. (E) The heterochromatin vs. euchromatin features of 10058-F4 (Traf3ip3) and insensitive genes (Hsd11b1 & Bex1).
Figure 7. Myc controls cellular subnetworks by transcriptionally varying their component concentrations
**Left--**Global up-regulation of MYC provokes linear and non-linear changes in outputs of cellular subsystems according to network architecture. By inducing activators and repressors, a panoply of feedback and coherent and incoherent feedforward loops are employed according to programs pre-existing in the cells. Right—Myc’s multiple partners have the potential to expedite passage through multiple stages of the transcription cycle and to create kinetic synergy (Chung and Levens, 2005; Herschlag and Johnson, 1993). Myc stimulated pause release and promoter reloading would amplify expression according to output levels.
Comment in
- All things to all people.
Littlewood TD, Kreuzaler P, Evan GI. Littlewood TD, et al. Cell. 2012 Sep 28;151(1):11-3. doi: 10.1016/j.cell.2012.09.006. Cell. 2012. PMID: 23021211 - Tumorigenesis: Megaphone MYC.
McCarthy N. McCarthy N. Nat Rev Cancer. 2012 Nov;12(11):733. doi: 10.1038/nrc3384. Epub 2012 Oct 5. Nat Rev Cancer. 2012. PMID: 23037452 No abstract available.
Similar articles
- Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells.
Kidder BL, Yang J, Palmer S. Kidder BL, et al. PLoS One. 2008;3(12):e3932. doi: 10.1371/journal.pone.0003932. Epub 2008 Dec 11. PLoS One. 2008. PMID: 19079543 Free PMC article. - Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis.
Sabò A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, Tonelli C, Fagà G, Bianchi V, Ronchi A, Low D, Müller H, Guccione E, Campaner S, Amati B. Sabò A, et al. Nature. 2014 Jul 24;511(7510):488-492. doi: 10.1038/nature13537. Epub 2014 Jul 9. Nature. 2014. PMID: 25043028 Free PMC article. - ZFP281 Recruits MYC to Active Promoters in Regulating Transcriptional Initiation and Elongation.
Luo Z, Liu X, Xie H, Wang Y, Lin C. Luo Z, et al. Mol Cell Biol. 2019 Nov 25;39(24):e00329-19. doi: 10.1128/MCB.00329-19. Print 2019 Dec 15. Mol Cell Biol. 2019. PMID: 31570506 Free PMC article. - Activation by c-Myc of transcription by RNA polymerases I, II and III.
Gomez-Roman N, Felton-Edkins ZA, Kenneth NS, Goodfellow SJ, Athineos D, Zhang J, Ramsbottom BA, Innes F, Kantidakis T, Kerr ER, Brodie J, Grandori C, White RJ. Gomez-Roman N, et al. Biochem Soc Symp. 2006;(73):141-54. doi: 10.1042/bss0730141. Biochem Soc Symp. 2006. PMID: 16626295 Review. - Acting locally and globally: Myc's ever-expanding roles on chromatin.
Varlakhanova NV, Knoepfler PS. Varlakhanova NV, et al. Cancer Res. 2009 Oct 1;69(19):7487-90. doi: 10.1158/0008-5472.CAN-08-4832. Epub 2009 Sep 22. Cancer Res. 2009. PMID: 19773445 Review.
Cited by
- A Cyclin D1-Dependent Transcriptional Program Predicts Clinical Outcome in Mantle Cell Lymphoma.
Demajo S, Albero R, Clot G, Castellano G, Navarro A, Capdevila C, Enjuanes A, Nadeu F, Giné E, Pinyol M, Jaffe ES, Ott G, Staudt LM, Rosenwald A, Scott DW, Rimsza LM, López-Guillermo A, Beà S, Campo E, Jares P. Demajo S, et al. Clin Cancer Res. 2021 Jan 1;27(1):213-225. doi: 10.1158/1078-0432.CCR-20-2868. Epub 2020 Oct 12. Clin Cancer Res. 2021. PMID: 33046520 Free PMC article. - Signal transduction in cancer.
Sever R, Brugge JS. Sever R, et al. Cold Spring Harb Perspect Med. 2015 Apr 1;5(4):a006098. doi: 10.1101/cshperspect.a006098. Cold Spring Harb Perspect Med. 2015. PMID: 25833940 Free PMC article. Review. - Revisiting global gene expression analysis.
Lovén J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. Lovén J, et al. Cell. 2012 Oct 26;151(3):476-82. doi: 10.1016/j.cell.2012.10.012. Cell. 2012. PMID: 23101621 Free PMC article. Review. - Regulation of miRNA biogenesis and turnover in the immune system.
Bronevetsky Y, Ansel KM. Bronevetsky Y, et al. Immunol Rev. 2013 May;253(1):304-16. doi: 10.1111/imr.12059. Immunol Rev. 2013. PMID: 23550654 Free PMC article. Review. - Different promoter affinities account for specificity in MYC-dependent gene regulation.
Lorenzin F, Benary U, Baluapuri A, Walz S, Jung LA, von Eyss B, Kisker C, Wolf J, Eilers M, Wolf E. Lorenzin F, et al. Elife. 2016 Jul 27;5:e15161. doi: 10.7554/eLife.15161. Elife. 2016. PMID: 27460974 Free PMC article.
References
- Alon U. An introduction to systems biology : design principles of biological circuits. Boca Raton, FL: Chapman & Hall/CRC; 2007.
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
- Berns K, Hijmans EM, Koh E, Daley GQ, Bernards R. A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene. 2000;19:3330–3334. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials