Purification, crystallization and X-ray diffraction analysis of human dynamin-related protein 1 GTPase-GED fusion protein - PubMed (original) (raw)
Purification, crystallization and X-ray diffraction analysis of human dynamin-related protein 1 GTPase-GED fusion protein
Eva Klinglmayr et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012.
Abstract
The mechano-enzyme dynamin-related protein 1 plays an important role in mitochondrial fission and is implicated in cell physiology. Dysregulation of Drp1 is associated with abnormal mitochondrial dynamics and neuronal damage. Drp1 shares structural and functional similarities with dynamin 1 with respect to domain organization, ability to self-assemble into spiral-like oligomers and GTP-cycle-dependent membrane scission. Structural studies of human dynamin-1 have greatly improved the understanding of this prototypical member of the dynamin superfamily. However, high-resolution structural information for full-length human Drp1 covering the GTPase domain, the middle domain and the GTPase effector domain (GED) is still lacking. In order to obtain mechanistic insights into the catalytic activity, a nucleotide-free GTPase-GED fusion protein of human Drp1 was expressed, purified and crystallized. Initial X-ray diffraction experiments yielded data to 2.67 Å resolution. The hexagonal-shaped crystals belonged to space group P2(1)2(1)2, with unit-cell parameters a = 53.59, b = 151.65, c = 43.53 Å, one molecule per asymmetric unit and a solvent content of 42%. Expression of selenomethionine-labelled protein is currently in progress. Here, the expression, purification, crystallization and X-ray diffraction analysis of the Drp1 GTPase-GED fusion protein are presented, which form a basis for more detailed structural and biophysical analysis.
Figures
Figure 1
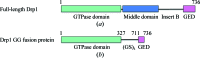
Construct design and schematic representation of the Drp1 GG fusion protein. (a) Full-length Drp1 consists of an N-terminal GTPase domain (green) and a helical middle domain (blue) followed by an unstructured region (insert B or variable domain) and a C-terminal GED (pink). (b) The GG fusion protein consists of the complete GTPase domain (amino acids 1–327) fused to a C-terminal fragment of the GED (amino acids 711–736) via a (GS)4 linker.
Figure 2
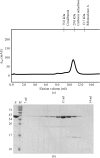
Purification of the Drp1 GG fusion protein. (a) Size-exclusion analysis. After Ni–NTA purification, proteins were loaded onto a Superdex 75 gel-filtration column and fractions containing protein were collected. In the absence of nucleotides, the protein eluted as a monomer at a retention volume of approximately 11 ml, which corresponds to a molecular mass of approximately 40 kDa. The retention volumes of molecular-mass standards (GE Healthcare) are displayed at the top. (b) SDS–PAGE analysis. Protein fractions were loaded onto a 15% SDS–PAGE gel and visualized by Coomassie Brilliant Blue staining. Lane E, eluted protein; lane M, protein molecular-mass markers (labelled in kDa on the left). The retention volumes are indicated at the top.
Figure 3
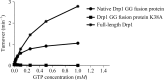
The Drp1 GG fusion protein displays GTPase activity: steady-state GTPase activities of Drp1 GG fusion protein (native Drp1 GG fusion protein) in comparison to Drp1 full-length isoform 2 and enzymatically inactive K38A mutant (Drp1 GG fusion protein K38A). Data are the mean ± standard deviation from three independent measurements.
Figure 4
Crystal forms of the Drp1 GG fusion protein. (a) In condition A only star-shaped intergrown crystals were present which could not be reproduced. (b) Reproducible crystals from condition B were only about 50 × 40 × 10 µm in size.
Figure 5
Diffraction image of the Drp1 GG fusion protein: a diffraction image of the crystals produced in condition B collected on BL14.1 at BESSY II, Berlin, Germany.
Similar articles
- Purification, crystallization and preliminary X-ray crystallographic analysis of Arabidopsis thaliana dynamin-related protein 1A GTPase-GED fusion protein.
Chen X, Xu X, Sun Y, Zhou J, Ma Y, Yan L, Lou Z. Chen X, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Jan 1;68(Pt 1):69-72. doi: 10.1107/S1744309111047634. Epub 2011 Dec 24. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 22232176 Free PMC article. - Intra- and intermolecular domain interactions of the C-terminal GTPase effector domain of the multimeric dynamin-like GTPase Drp1.
Zhu PP, Patterson A, Stadler J, Seeburg DP, Sheng M, Blackstone C. Zhu PP, et al. J Biol Chem. 2004 Aug 20;279(34):35967-74. doi: 10.1074/jbc.M404105200. Epub 2004 Jun 18. J Biol Chem. 2004. PMID: 15208300 - Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology.
Chang CR, Blackstone C. Chang CR, et al. J Biol Chem. 2007 Jul 27;282(30):21583-7. doi: 10.1074/jbc.C700083200. Epub 2007 Jun 6. J Biol Chem. 2007. PMID: 17553808 - New insights into the function and regulation of mitochondrial fission.
Otera H, Ishihara N, Mihara K. Otera H, et al. Biochim Biophys Acta. 2013 May;1833(5):1256-68. doi: 10.1016/j.bbamcr.2013.02.002. Epub 2013 Feb 20. Biochim Biophys Acta. 2013. PMID: 23434681 Review. - Dynamin and its role in membrane fission.
Hinshaw JE. Hinshaw JE. Annu Rev Cell Dev Biol. 2000;16:483-519. doi: 10.1146/annurev.cellbio.16.1.483. Annu Rev Cell Dev Biol. 2000. PMID: 11031245 Free PMC article. Review.
Cited by
- Functional mapping of human dynamin-1-like GTPase domain based on x-ray structure analyses.
Wenger J, Klinglmayr E, Fröhlich C, Eibl C, Gimeno A, Hessenberger M, Puehringer S, Daumke O, Goettig P. Wenger J, et al. PLoS One. 2013 Aug 19;8(8):e71835. doi: 10.1371/journal.pone.0071835. eCollection 2013. PLoS One. 2013. PMID: 23977156 Free PMC article. - Steric interference from intrinsically disordered regions controls dynamin-related protein 1 self-assembly during mitochondrial fission.
Lu B, Kennedy B, Clinton RW, Wang EJ, McHugh D, Stepanyants N, Macdonald PJ, Mears JA, Qi X, Ramachandran R. Lu B, et al. Sci Rep. 2018 Jul 18;8(1):10879. doi: 10.1038/s41598-018-29001-9. Sci Rep. 2018. PMID: 30022112 Free PMC article.
References
- Bereiter-Hahn, J. & Vöth, M. (1994). Microsc. Res. Tech. 27, 198–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- J 3173/FWF_/Austrian Science Fund FWF/Austria
- R01 EY016164/EY/NEI NIH HHS/United States
- R01 NS055193/NS/NINDS NIH HHS/United States
- R01 NS055195/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous