Genomics of adaptation during experimental evolution of the opportunistic pathogen Pseudomonas aeruginosa - PubMed (original) (raw)
Genomics of adaptation during experimental evolution of the opportunistic pathogen Pseudomonas aeruginosa
Alex Wong et al. PLoS Genet. 2012 Sep.
Abstract
Adaptation is likely to be an important determinant of the success of many pathogens, for example when colonizing a new host species, when challenged by antibiotic treatment, or in governing the establishment and progress of long-term chronic infection. Yet, the genomic basis of adaptation is poorly understood in general, and for pathogens in particular. We investigated the genetics of adaptation to cystic fibrosis-like culture conditions in the presence and absence of fluoroquinolone antibiotics using the opportunistic pathogen Pseudomonas aeruginosa. Whole-genome sequencing of experimentally evolved isolates revealed parallel evolution at a handful of known antibiotic resistance genes. While the level of antibiotic resistance was largely determined by these known resistance genes, the costs of resistance were instead attributable to a number of mutations that were specific to individual experimental isolates. Notably, stereotypical quinolone resistance mutations in DNA gyrase often co-occurred with other mutations that, together, conferred high levels of resistance but no consistent cost of resistance. This result may explain why these mutations are so prevalent in clinical quinolone-resistant isolates. In addition, genes involved in cyclic-di-GMP signalling were repeatedly mutated in populations evolved in viscous culture media, suggesting a shared mechanism of adaptation to this CF-like growth environment. Experimental evolutionary approaches to understanding pathogen adaptation should provide an important complement to studies of the evolution of clinical isolates.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Minimum inhibitory concentration (MIC) to ciprofloxacin for each of 48 experimentally evolved populations (“pop”) or single gentoypes (“single”).
Populations were evolved in the presence (black) or absence (grey) of 1 µg/ml ciprofloxacin for 8 days (∼50 generations). Two different media were used, as indicated on the X-axis.
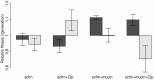
Figure 2. Competitive fitness of experimentally evolved populations (dark bars) or single genotypes (light bars).
Fitness was measured in the absence of antibiotic via direct competitions with a lacZ marked ancestral strain (Pa14). The height of each bar indicates mean fitness for 12 evolved populations (genotypes), with the error bar giving +/−1 SE. Competitions were carried out in scfm for populations (genotypes) evolved in scfm or in scfm+ciprofloxacin, and in scfm+mucin for populations (genotypes) evolved in scfm+mucin or in scfm+mucin+ciprofloxacin. Fitness below one indicates low fitness relative to the ancestor, while fitness above one indicates an overall benefit in the absence of antibiotic.
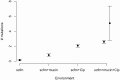
Figure 3. Numbers of mutations identified in evolved genotypes.
Mean number of mutations by treatment, with error bars giving +/−1 SE. In the “scfm+mucin+Cip” treatment, the filled circle represents all evolved genotypes, and the filled diamond represents the ten non-mutator genotypes.
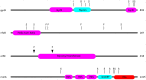
Figure 4. Locations of mutations in the ciprofloxacin resistance proteins nfxB, orfN, gyrB, and in the putatively mucin-adaptive protein morA.
“S”: location of a single nucleotide polymorphism; “F”: frameshift; “I”: in-frame insertion; “D”: in-frame deletion.
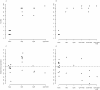
Figure 5. Mutations in known resistance genes are strong predictors of ciprofloxacin resistance, but not fitness.
Fold-increase in MIC (A and B), or relative fitness (C and D), for genotypes bearing the given mutations, with genotypes evolved in scfm (A and C) or scfm+mucin (B and D).
Figure 6. Variation in colony morphology.
Isolates were grown on 1% tryptone plates containing Congo red and Coomassie blue. (A) Ancestral Pa14; (B) strain smA1 carrying a single mutation in morA (H975Y); (C) strain smA2 carrying a different mutation in morA (L1155Q); (D) strain smD6 bearing three mutations, including one in wspF (an out-of-frame deletion); (E) strain smC3 bearing a single mutation in Pa14_56280 (M204I); (F) strain scfmB5 carrying a single mutation in nfxB (G180S).
Similar articles
- Evolved Aztreonam Resistance Is Multifactorial and Can Produce Hypervirulence in Pseudomonas aeruginosa.
Jorth P, McLean K, Ratjen A, Secor PR, Bautista GE, Ravishankar S, Rezayat A, Garudathri J, Harrison JJ, Harwood RA, Penewit K, Waalkes A, Singh PK, Salipante SJ. Jorth P, et al. mBio. 2017 Oct 31;8(5):e00517-17. doi: 10.1128/mBio.00517-17. mBio. 2017. PMID: 29089424 Free PMC article. - Evolutionary genomics of epidemic and nonepidemic strains of Pseudomonas aeruginosa.
Dettman JR, Rodrigue N, Aaron SD, Kassen R. Dettman JR, et al. Proc Natl Acad Sci U S A. 2013 Dec 24;110(52):21065-70. doi: 10.1073/pnas.1307862110. Epub 2013 Dec 9. Proc Natl Acad Sci U S A. 2013. PMID: 24324153 Free PMC article. - Evolution of Antibiotic Resistance in Biofilm and Planktonic Pseudomonas aeruginosa Populations Exposed to Subinhibitory Levels of Ciprofloxacin.
Ahmed MN, Porse A, Sommer MOA, Høiby N, Ciofu O. Ahmed MN, et al. Antimicrob Agents Chemother. 2018 Jul 27;62(8):e00320-18. doi: 10.1128/AAC.00320-18. Print 2018 Aug. Antimicrob Agents Chemother. 2018. PMID: 29760140 Free PMC article. - Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: new approaches to an old problem.
Rehman A, Patrick WM, Lamont IL. Rehman A, et al. J Med Microbiol. 2019 Jan;68(1):1-10. doi: 10.1099/jmm.0.000873. J Med Microbiol. 2019. PMID: 30605076 Review. - Parallel evolution and local differentiation in quinolone resistance in Pseudomonas aeruginosa.
Wong A, Kassen R. Wong A, et al. Microbiology (Reading). 2011 Apr;157(Pt 4):937-944. doi: 10.1099/mic.0.046870-0. Epub 2011 Feb 3. Microbiology (Reading). 2011. PMID: 21292748 Review.
Cited by
- Genomic analysis of parallel-evolved cyanobacterium Synechocystis sp. PCC 6803 under acid stress.
Uchiyama J, Kanesaki Y, Iwata N, Asakura R, Funamizu K, Tasaki R, Agatsuma M, Tahara H, Matsuhashi A, Yoshikawa H, Ogawa S, Ohta H. Uchiyama J, et al. Photosynth Res. 2015 Aug;125(1-2):243-54. doi: 10.1007/s11120-015-0111-3. Epub 2015 Mar 4. Photosynth Res. 2015. PMID: 25736465 - Biological lessons for strategic resistance management.
Blumstein DT, Johnson NA, Katz ND, Kharpatin S, Ortiz-Ross X, Parra E, Reshke A. Blumstein DT, et al. Evol Appl. 2023 Nov 15;16(12):1861-1871. doi: 10.1111/eva.13616. eCollection 2023 Dec. Evol Appl. 2023. PMID: 38143901 Free PMC article. - Sublethal ciprofloxacin treatment leads to rapid development of high-level ciprofloxacin resistance during long-term experimental evolution of Pseudomonas aeruginosa.
Jørgensen KM, Wassermann T, Jensen PØ, Hengzuang W, Molin S, Høiby N, Ciofu O. Jørgensen KM, et al. Antimicrob Agents Chemother. 2013 Sep;57(9):4215-21. doi: 10.1128/AAC.00493-13. Epub 2013 Jun 17. Antimicrob Agents Chemother. 2013. PMID: 23774442 Free PMC article. - Biofilm Maintenance as an Active Process: Evidence that Biofilms Work Hard to Stay Put.
Katharios-Lanwermeyer S, O'Toole GA. Katharios-Lanwermeyer S, et al. J Bacteriol. 2022 Apr 19;204(4):e0058721. doi: 10.1128/jb.00587-21. Epub 2022 Mar 21. J Bacteriol. 2022. PMID: 35311557 Free PMC article. Review. - Mutation-Driven Evolution of Pseudomonas aeruginosa in the Presence of either Ceftazidime or Ceftazidime-Avibactam.
Sanz-García F, Hernando-Amado S, Martínez JL. Sanz-García F, et al. Antimicrob Agents Chemother. 2018 Sep 24;62(10):e01379-18. doi: 10.1128/AAC.01379-18. Print 2018 Oct. Antimicrob Agents Chemother. 2018. PMID: 30082283 Free PMC article.
References
- Pasteur L (1878) La Theorie des Germes. Comptes Rendus l'Academie des Sciences 86: 1037–1043.
- Garber ED (1960) The host as a growth medium. Ann NY Acad Sci 88: 1187–1194. - PubMed
- Andersson DI (2003) Persistence of antibiotic resistant bacteria. Curr Opin Microbiol l6: 452–456. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources