The Kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart - PubMed (original) (raw)
The Kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart
Lisa Korostowski et al. PLoS Genet. 2012 Sep.
Abstract
Although many of the questions raised by the discovery of imprinting have been answered, we have not yet accounted for tissue- or stage-specific imprinting. The Kcnq1 imprinted domain exhibits complex tissue-specific expression patterns co-existing with a domain-wide cis-acting control element. Transcription of the paternally expressed antisense non-coding RNA Kcnq1ot1 silences some neighboring genes in the embryo, while others are unaffected. Kcnq1 is imprinted in early cardiac development but becomes biallelic after midgestation. To explore this phenomenon and the role of Kcnq1ot1, we used allele-specific assays and chromosome conformational studies in wild-type mice and mice with a premature termination mutation for Kcnq1ot1. We show that Kcnq1 imprinting in early heart is established and maintained independently of Kcnq1ot1 expression, thus excluding a role for Kcnq1ot1 in repressing Kcnq1, even while silencing other genes in the domain. The exact timing of the mono- to biallelic transition is strain-dependent, with the CAST/EiJ allele becoming activated earlier and acquiring higher levels than the C57BL/6J allele. Unexpectedly, Kcnq1ot1 itself also switches to biallelic expression specifically in the heart, suggesting that tissue-specific loss of imprinting may be common during embryogenesis. The maternal Kcnq1ot1 transcript is shorter than the paternal ncRNA, and its activation depends on an alternative transcriptional start site that bypasses the maternally methylated promoter. Production of Kcnq1ot1 on the maternal chromosome does not silence Cdkn1c. We find that in later developmental stages, however, Kcnq1ot1 has a role in modulating Kcnq1 levels, since its absence leads to overexpression of Kcnq1, an event accompanied by an aberrant three-dimensional structure of the chromatin. Thus, our studies reveal regulatory mechanisms within the Kcnq1 imprinted domain that operate exclusively in the heart on Kcnq1, a gene crucial for heart development and function. We also uncover a novel mechanism by which an antisense non-coding RNA affects transcription through regulating chromatin flexibility and access to enhancers.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
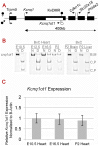
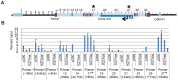
Figure 1. Cardiac expression profile of Kcnq1ot1.
A) Imprinting pattern in the Kcnq1 domain in early development in the embryo. Arrows above the line represent maternal transcription and those below represent paternal transcription. B) Kcnq1ot1 imprinting as determined by RT-PCR followed by allele-specific restriction digest. The maternal Kcnq1ot1 becomes activated as development progresses in the heart. However, in brain and liver, Kcnq1ot1 remains monoallelic. M, maternal; P, paternal; N, non-digested; D, Digested; B, C57BL/6J; C, B6(CAST7). C) Quantitative analysis by qRT-PCR of Kcnq1ot1 expression throughout development. RNA levels were normalized to β-actin.
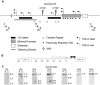
Figure 2. Characterization of the maternal Kcnq1ot1 transcript.
A) Schematic showing the regulatory sequences at the Kcnq1ot1 locus. Minimal promoter, enhancer and previously reported transcriptional start sites (*, TSS) are from Fitzpatrick et al. (. The silencing domain depicted was characterized by Mohammad et al. (. Primers used for 5′ RACE experiments in this report are designated A, B, C and nested primers, An, Bn and Cn. Transcriptional start sites (TSS) in heart and liver are depicted as bent arrows. A star indicates the relative location of a single nucleotide polymorphism (SNP) used to discriminate between parental transcripts. B) Schematic of the scan to determine the length of the Kcnq1ot1 maternal transcript, with primers amplifying fragments located at the indicated distances relative to the Kcnq1ot1 transcriptional start site. C) RT-PCRs on RNAs extracted from hearts of F1 hybrid progeny from a BxC cross, followed by allele-specific digestion, showing the maternal transcript absent after 33 kb. M, maternal; P, paternal; ND, non-digested; D, digested; B, C57BL/6J; C, B6(CAST7).
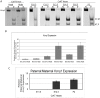
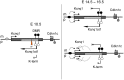
Figure 3. Kcnq1 expression in the heart during development when the truncated Kcnq1ot1 (K-term mutation) is inherited paternally.
A) RT-PCR followed by allele-specific digests in E10.5 heads and bodies and throughout the development of the heart in F1 hybrid progeny of B6(CAST7)×K-term mice. M, maternal; P, paternal; N, non-digested; D, digested; B, C57BL/6J; KT, K-term. B) qRT-PCR analysis of Kcnq1 expression in wild-type and K-term mice. Transcripts were normalized to β-actin. A significant difference in expression was seen when comparing wild-type and K-term hearts at E16.5 and P2 Hearts. These differences had a p-value less than 0.05. C) Parental origin of Kcnq1 expression throughout cardiac development. The RT-PCR and allele specific bands were quantified and the ratio of paternal to maternal transcript was determined.
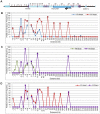
Figure 4. Chromosome conformation capture (3C) in the K-term mouse.
A) Schematic of the Kcnq1 domain. The blue arrowhead indicates the anchor primer at the Kcnq1 promoter and vertical lines represent the regions investigated. The purple boxes represent Kcnq1 exons, the dark blue box represents the Kcnq1ot1 gene. Comparison of the chromatin interaction profile in wild-type and K-term hearts (B), wild-type and K-term brains (C) and K-term hearts and brains (D).
Figure 5. Chromatin immunoprecipitation (ChIP) of selected regions of the Kcnq1 gene.
A) Schematic of the Kcnq1 domain. The blue arrowhead indicates the anchor primer at the Kcnq1 promoter and vertical lines represents the primer regions investigated. The purple boxes represent Kcnq1 exons, the dark blue box represents the Kcnq1ot1 gene. B) ChIP analysis in wild-type heart for p300, H3K4Me1 and H3K27Ac at the interaction peaks observed in the 3C assays. The asterisks indicate frequent interactions in the K-term heart as determined by 3C.
Figure 6. Model for regulation of Kcnq1 in the embryonic heart.
WT, wild-type; K-term, mutant mouse, in which transcription of Kcnq1ot1 is terminated prematurely; IF, methylation sensitive inhibitory factor. Maternal (m) events are shown above and paternal (p) events below the chromosome; filled circles, methylated DNA, empty circles, unmethylated DNA. Curved arrows represent interactions, bent arrows depict transcription. Ovals represent enhancers, which are inactive (light gray) at 10.5 dpc and active (black) at 14.5–16.5 dpc.
Similar articles
- Enhancers compete with a long non-coding RNA for regulation of the Kcnq1 domain.
Schultz BM, Gallicio GA, Cesaroni M, Lupey LN, Engel N. Schultz BM, et al. Nucleic Acids Res. 2015 Jan;43(2):745-59. doi: 10.1093/nar/gku1324. Epub 2014 Dec 24. Nucleic Acids Res. 2015. PMID: 25539921 Free PMC article. - Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus.
Zhang H, Zeitz MJ, Wang H, Niu B, Ge S, Li W, Cui J, Wang G, Qian G, Higgins MJ, Fan X, Hoffman AR, Hu JF. Zhang H, et al. J Cell Biol. 2014 Jan 6;204(1):61-75. doi: 10.1083/jcb.201304152. J Cell Biol. 2014. PMID: 24395636 Free PMC article. - Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo.
Lewis A, Green K, Dawson C, Redrup L, Huynh KD, Lee JT, Hemberger M, Reik W. Lewis A, et al. Development. 2006 Nov;133(21):4203-10. doi: 10.1242/dev.02612. Epub 2006 Oct 4. Development. 2006. PMID: 17021040 - Kcnq1ot1: a chromatin regulatory RNA.
Kanduri C. Kanduri C. Semin Cell Dev Biol. 2011 Jun;22(4):343-50. doi: 10.1016/j.semcdb.2011.02.020. Epub 2011 Feb 21. Semin Cell Dev Biol. 2011. PMID: 21345374 Review. - Epigenetics of imprinted long noncoding RNAs.
Mohammad F, Mondal T, Kanduri C. Mohammad F, et al. Epigenetics. 2009 Jul 1;4(5):277-86. Epub 2009 Jul 10. Epigenetics. 2009. PMID: 19617707 Review.
Cited by
- Enhancers compete with a long non-coding RNA for regulation of the Kcnq1 domain.
Schultz BM, Gallicio GA, Cesaroni M, Lupey LN, Engel N. Schultz BM, et al. Nucleic Acids Res. 2015 Jan;43(2):745-59. doi: 10.1093/nar/gku1324. Epub 2014 Dec 24. Nucleic Acids Res. 2015. PMID: 25539921 Free PMC article. - Parental bias in expression and interaction of genes in the equine placenta.
Dini P, Kalbfleisch T, Uribe-Salazar JM, Carossino M, Ali HE, Loux SC, Esteller-Vico A, Norris JK, Anand L, Scoggin KE, Rodriguez Lopez CM, Breen J, Bailey E, Daels P, Ball BA. Dini P, et al. Proc Natl Acad Sci U S A. 2021 Apr 20;118(16):e2006474118. doi: 10.1073/pnas.2006474118. Proc Natl Acad Sci U S A. 2021. PMID: 33853939 Free PMC article. - Epigenetic and transcriptional features of the novel human imprinted lncRNA GPR1AS suggest it is a functional ortholog to mouse Zdbf2linc.
Kobayashi H, Yanagisawa E, Sakashita A, Sugawara N, Kumakura S, Ogawa H, Akutsu H, Hata K, Nakabayashi K, Kono T. Kobayashi H, et al. Epigenetics. 2013 Jun;8(6):635-45. doi: 10.4161/epi.24887. Epub 2013 May 9. Epigenetics. 2013. PMID: 23764515 Free PMC article. - Comparison of Long Non-Coding RNA Expression Profiles of Cattle and Buffalo Differing in Muscle Characteristics.
Li H, Huang K, Wang P, Feng T, Shi D, Cui K, Luo C, Shafique L, Qian Q, Ruan J, Liu Q. Li H, et al. Front Genet. 2020 Feb 26;11:98. doi: 10.3389/fgene.2020.00098. eCollection 2020. Front Genet. 2020. PMID: 32174968 Free PMC article. - Comprehensive Analysis of mRNA, lncRNA, circRNA, and miRNA Expression Profiles and Their ceRNA Networks in the Longissimus Dorsi Muscle of Cattle-Yak and Yak.
Huang C, Ge F, Ma X, Dai R, Dingkao R, Zhaxi Z, Burenchao G, Bao P, Wu X, Guo X, Chu M, Yan P, Liang C. Huang C, et al. Front Genet. 2021 Dec 13;12:772557. doi: 10.3389/fgene.2021.772557. eCollection 2021. Front Genet. 2021. PMID: 34966412 Free PMC article.
References
- Sleutels F, Zwart R, Barlow DP (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810–813. - PubMed
- Borsoni G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, et al. (1991) Characterization of a murine gene expressed from the inactive X chromosome. Nature 351: 325–328. - PubMed
- Lee JT, Lu N (1999) Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99: 47–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K22 CA140361/CA/NCI NIH HHS/United States
- R01 GM093066/GM/NIGMS NIH HHS/United States
- K22CA140361-01/CA/NCI NIH HHS/United States
- R01GM093066/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous