Activation-induced cytidine deaminase alters the subcellular localization of Tet family proteins - PubMed (original) (raw)
Activation-induced cytidine deaminase alters the subcellular localization of Tet family proteins
Yuko Arioka et al. PLoS One. 2012.
Abstract
Activation-induced cytidine deminase (Aid), a unique enzyme that deaminates cytosine in DNA, shuttles between the nucleus and the cytoplasm. A recent study proposed a novel function of Aid in active DNA demethylation via deamination of 5-hydroxymethylcytosine, which is converted from 5-methylcytosine by the Ten-eleven translocation (Tet) family of enzymes. In this study, we examined the effect of simultaneous expression of Aid and Tet family proteins on the subcellular localization of each protein. We found that overexpressed Aid is mainly localized in the cytoplasm, whereas Tet1 and Tet2 are localized in the nucleus, and Tet3 is localized in both the cytoplasm and the nucleus. However, nuclear Tet proteins were gradually translocated to the cytoplasm when co-expressed with Aid. We also show that Aid-mediated translocation of Tet proteins is associated with Aid shuttling. Here we propose a possible role for Aid as a regulator of the subcellular localization of Tet family proteins.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
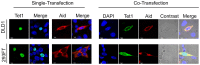
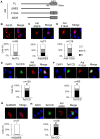
Figure 1. Overexpressed Aid alters the subcellular localization of Tet1.
Images of cells transiently expressing N-terminally Xpress-tagged Tet1 or C-terminally Myc-tagged Aid. Tet1 was predominantly localized in the nucleus, whereas Aid was mainly localized in the cytoplasm 48 h after transfection in both DLD-1 and HEK293FT cells. When cells were co-transfected with a plasmid expressing Aid, the Tet1 subcellular localization was altered to the cytoplasm, where Aid was mainly localized. The scale bar is 10 µm.
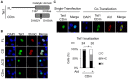
Figure 2. Tet1 translocation requires its catalytic domain.
(A) A schematic representation of the Tet1 structure and its mutants used in this study. (aa = amino acid). (B) Confocal images of DLD-1 cells transiently expressing N-terminally Xpress-tagged Tet1 mutants with or without C-terminally Myc-tagged Aid. All Tet1 constructs (FL, CD and ΔCD) were localized in the nucleus when solely expressed in DLD-1 cells. When co-expressed with Aid, Tet1FL and Tet1CD were translocated to the cytoplasm, whereas Tet1ΔCD remained in the nucleus. (C) Each bar represents the proportion of cells with the different localizations of Tet1. The number (n) of cells indicated above each bar was scored according to their subcellular localization. N (black); nuclear localization, N+C (gray); both nuclear and cytoplasmic localization, C (white); cytoplasmic localization in multiple microscope fields. The scale bars in images are 10 µm. *p<0.01.
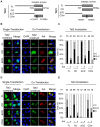
Figure 3. Tet1 translocation in the presence of Aid is independent of the Tet1 enzymatic activity.
(A) A schematic representation of the Tet1CD mutant (CDm) used in this study. Tet1CDm were tagged with N-terminal Xpress. (B) Tet1FL and CD had enzyme activity and produced 5hmC, but Tet1ΔCD and CDm did not. (C) C-terminally Myc-tagged Aid expression altered the subcellular localization of Tet1CDm, which lacks the enzymatic activity. The upper panels are representative images of DLD-1 cells transiently expressing Tet1CDm with or without simultaneous expression of Aid. The lower graph shows the percentage score of the examined transfected cells (indicated as a number). The scale bars are 10 µm. *p<0.01. N (black); nuclear localization, N+C (gray); both nuclear and cytoplasmic localization, C (white); cytoplasmic localization in multiple microscope fields.
Figure 4. The subcellular localization of Tet2 and Tet3 is altered by Aid expression.
(A) A schematic representation of the Tet2 and Tet3 structures and their mutants used in this study. (B) N-terminally Xpress-tagged Tet2 or its mutants with or without Aid tagged with C-terminal Myc were imaged by confocal microscopy in transiently transfected DLD-1 cells. (C) The number (n) of cells indicated above each bar was scored according to Tet2 subcellular localization. All Tet2 mutants were translocated to the cytoplasm in the presence of Aid (p<0.01, vs in the absence of Aid). (D, E) Simultaneous expression of N-terminally Xpress-tagged Tet3 and Aid-Myc. Tet3FL, CD and CDm were translocated to the cytoplasm when co-expressed with Aid (p<0.01, single-expression vs co-expression). Tet3ΔCD was localized in the cytoplasm regardless of the Aid expression. The scale bars are 10 µm. N (black); nuclear localization, N+C (gray); both nuclear and cytoplasmic localization, C (white); cytoplasmic localization in multiple microscope fields.
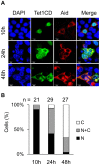
Figure 5. Nuclear Tet1 is gradually translocated into the cytoplasm by the simultaneous expression of Aid.
(A) Confocal images of HEK293FT cells transiently co-expressing Tet1CD tagged with N-terminal Xpress and Aid tagged with C-terminal Myc at different time points (10 h, 24 h and 48 h) after co-transfection. The scale bars in images are 10 µm. (B) The number (n) of cells indicated above each bar was scored according to the Tet1CD subcellular localization. The number of cells with cytoplasmic Tet1CD gradually increased after co-transfection. N (black); nuclear localization, N+C (gray); both nuclear and cytoplasmic localization, C (white); cytoplasmic localization in multiple microscope fields.
Figure 6. Aid shuttling is associated with Aid-mediating transcloation of Tet1.
(A) A schematic representation of the Aid structure and its mutants used in this study. All Aid constructs were tagged with C-terminal Myc. (B–D) The upper figures are representative confocal images of DLD-1 cells transiently expressing only Aid FL (B), ΔNES (C) or F193A (D). The lower figure represents the proportion of cells with different subcellular localization of Aid. Aid mutants defect in NES showed the increased nuclear localization. *p<0.05 vs Aid FL. (E, F) The upper figures are representative confocal images of DLD-1 cells transiently co-expressing AidΔNES and Tet1CD (E), or AidF193A and Tet1CD (F). Tet1CD were tagged with N-terminal Xpress. The lower figure shows the proportion of cells with different localizations of Tet1 (E; AidΔNES and Tet1CD, F; AidF193A and Tet1CD). Aid mutants, which exhibit impaired shuttling between the nucleus and the cytoplasm, failed to alter the subcellular localization of Tet1. #p<0.05 vs with Aid FL. (G) The upper figures are representative confocal images of DLD-1 cells transiently expressing only AidΔN26. The lower figure represents the proportion of cells with different subcellular localization of AidΔN26. *p<0.05 vs Aid FL. (H) The upper figures are representative confocal images of DLD-1 cells transiently co-expressing AidΔN26 and Tet1CD. The lower figure shows the proportion of cells with different localizations of Tet1. #p<0.05 vs with AidFL. The scale bars are 10 µm. N (black); nuclear localization, N+C (gray); both nuclear and cytoplasmic localization, C (white); cytoplasmic localization in multiple microscope fields.
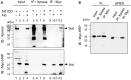
Figure 7. Aid interacts with Tet1CD.
(A) Tet1CD was co-immunoprecipitated with Aid FL. Lysates from HEK293FT cells transfected with N-terminally Xpress-tagged Tet1CD, C-terminally Myc-tagged AidFL or both of them were immunoprecipitated (IP) by anti-Xpress mAbs or anti-Myc mAbs. Immunoblotting (IB) was performed by using an anti-Xpress Abs or anti-Myc-HRP antibody. Lane nos. 1, 5 and 9 were single transfections of Tet1CD. Lane nos. 2, 6 and 10 were single-transfections of Aid. Lane nos. 3, 7 and 11 were for mock transfection. Lane nos. 4, 8 and 12 shows the results for the co-transfection of Tet1CD and Aid. (B) The Co-IP experiment was performed by using lysates from HEK293FT cells co-transfected with N-terminally Xpress tagged-Tet1CD and C-terminally Myc-tagged Aid FL, or with N-terminally Xpress-tagged Tet1CD and C-terminally Myc-tagged AidΔNES. Despite the similar localization of AidΔNES and Tet1CD in the nucleus, AidΔNES revealed a decreased association with Tet1CD compared to Aid FL.
Similar articles
- TET enzymes augment activation-induced deaminase (AID) expression via 5-hydroxymethylcytosine modifications at the Aicda superenhancer.
Lio CJ, Shukla V, Samaniego-Castruita D, González-Avalos E, Chakraborty A, Yue X, Schatz DG, Ay F, Rao A. Lio CJ, et al. Sci Immunol. 2019 Apr 26;4(34):eaau7523. doi: 10.1126/sciimmunol.aau7523. Sci Immunol. 2019. PMID: 31028100 Free PMC article. - Tet-mediated formation of 5-hydroxymethylcytosine in RNA.
Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, Cai Q, Ji D, Jin SG, Niedernhofer LJ, Pfeifer GP, Xu GL, Wang Y. Fu L, et al. J Am Chem Soc. 2014 Aug 20;136(33):11582-5. doi: 10.1021/ja505305z. Epub 2014 Aug 7. J Am Chem Soc. 2014. PMID: 25073028 Free PMC article. - Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked β-N-acetylglucosamine transferase (OGT).
Zhang Q, Liu X, Gao W, Li P, Hou J, Li J, Wong J. Zhang Q, et al. J Biol Chem. 2014 Feb 28;289(9):5986-96. doi: 10.1074/jbc.M113.524140. Epub 2014 Jan 6. J Biol Chem. 2014. PMID: 24394411 Free PMC article. - Epigenetic Function of TET Family, 5-Methylcytosine, and 5-Hydroxymethylcytosine in Hematologic Malignancies.
Li W, Xu L. Li W, et al. Oncol Res Treat. 2019;42(6):309-318. doi: 10.1159/000498947. Epub 2019 May 3. Oncol Res Treat. 2019. PMID: 31055566 Review. - Mechanisms of TET protein-mediated DNA demethylation and its role in the regulation of mouse development.
Jia ZW, Gao SX, Zhang YC, Zhang XH. Jia ZW, et al. Yi Chuan. 2015 Jan;37(1):34-40. doi: 10.16288/j.yczz.2015.01.005. Yi Chuan. 2015. PMID: 25608811 Review.
Cited by
- Tracing TET1 expression in prostate cancer: discovery of malignant cells with a distinct oncogenic signature.
Schagdarsurengin U, Luo C, Slanina H, Sheridan D, Füssel S, Böğürcü-Seidel N, Gattenloehner S, Baretton GB, Hofbauer LC, Wagenlehner F, Dansranjav T. Schagdarsurengin U, et al. Clin Epigenetics. 2021 Nov 29;13(1):211. doi: 10.1186/s13148-021-01201-7. Clin Epigenetics. 2021. PMID: 34844636 Free PMC article. - A B-cell targeting virus disrupts potentially protective genomic methylation patterns in lymphoid tissue by increasing global 5-hydroxymethylcytosine levels.
Ciccone NA, Mwangi W, Ruzov A, Smith LP, Butter C, Nair V. Ciccone NA, et al. Vet Res. 2014 Oct 23;45(1):108. doi: 10.1186/s13567-014-0108-5. Vet Res. 2014. PMID: 25338704 Free PMC article. - Cell cycle and p53 gate the direct conversion of human fibroblasts to dopaminergic neurons.
Jiang H, Xu Z, Zhong P, Ren Y, Liang G, Schilling HA, Hu Z, Zhang Y, Wang X, Chen S, Yan Z, Feng J. Jiang H, et al. Nat Commun. 2015 Dec 7;6:10100. doi: 10.1038/ncomms10100. Nat Commun. 2015. PMID: 26639555 Free PMC article. - SIRT1 Activation Disrupts Maintenance of Myelodysplastic Syndrome Stem and Progenitor Cells by Restoring TET2 Function.
Sun J, He X, Zhu Y, Ding Z, Dong H, Feng Y, Du J, Wang H, Wu X, Zhang L, Yu X, Lin A, McDonald T, Zhao D, Wu H, Hua WK, Zhang B, Feng L, Tohyama K, Bhatia R, Oberdoerffer P, Chung YJ, Aplan PD, Boultwood J, Pellagatti A, Khaled S, Kortylewski M, Pichiorri F, Kuo YH, Carlesso N, Marcucci G, Jin H, Li L. Sun J, et al. Cell Stem Cell. 2018 Sep 6;23(3):355-369.e9. doi: 10.1016/j.stem.2018.07.018. Epub 2018 Aug 23. Cell Stem Cell. 2018. PMID: 30146412 Free PMC article. - The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases.
Solary E, Bernard OA, Tefferi A, Fuks F, Vainchenker W. Solary E, et al. Leukemia. 2014 Mar;28(3):485-96. doi: 10.1038/leu.2013.337. Epub 2013 Nov 13. Leukemia. 2014. PMID: 24220273 Review.
References
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21. - PubMed
- Goll MG, Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514. - PubMed
- Chen T, Li E (2004) Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Biol 60: 55–89. - PubMed
- Reik W, Dean W, Walter J (2001) Epigenetic reprogramming in mammalian development. Science 293: 1089–1093. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was supported by Grants-in-Aid from the Ministry of Health, Labour and Welfare of Japan, by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 23130508 and 24390096), and Grant from the Naito Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.