The histone variant macroH2A1.1 is recruited to DSBs through a mechanism involving PARP1 - PubMed (original) (raw)
The histone variant macroH2A1.1 is recruited to DSBs through a mechanism involving PARP1
Chang Xu et al. FEBS Lett. 2012.
Abstract
The repair of DNA double-strand breaks (DSBs) requires remodeling of the local chromatin architecture to allow the repair machinery to access sites of damage. Here, we report that the histone variant macroH2A1.1 is recruited to DSBs. Cells lacking macroH2A1 have defective recruitment of 53BP1, defective activation of chk2 kinase and increased radiosensitivity. Importantly, macroH2A1.1 is not incorporated into nucleosomes at DSBs, but instead associates with the chromatin through a mechanism which requires PARP1 activity. These results reveal an unusual mechanism involving a direct association of macroH2A1.1 with PARylated chromatin which is critical for retaining 53BP1 at sites of damage.
Copyright © 2012 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures
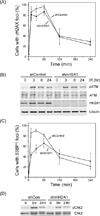
Figure 1. Depletion of macroH2A1 increases radiosensitivity
(A) 293T cells were infected with a non-specific shRNA (shCon) or shRNA targeting mH2A1.1 or mH2A1.2 (shmH2A1). mH2A1 was measured by western blot. (B) mH2A1.1 and mH2A1.2 mRNA levels were measured by RT-qPCR in shCon and shmH2A1 cells. (C) and (D) 293T cells expressing a non-specific shRNA (shCon: ●) or shmH2A1 (○) were irradiated (C) or incubated with MMS for 1 hr (D). Clonogenic cell survival was measured 10 days later. Results ± SE (n = 3). (E) HeLa cells with an integrated NHEJ-GFP reporter were stably infected with control shRNA or shRNA targeting mH2A1. I-SceI was transiently expressed and GFP+ cells counted by FACS 48hr later. Results ± SD (n = 3).
Figure 2. mH2A1 is required for recruitment of 53BP1 to DSBs
(A) Cells expressing control shRNA or shRNA targeting mH2A1 were irradiated (2Gy), fixed and processed for immunofluorescent staining using γH2AX antibody. The number of cells with >5 foci were counted. Results ± SD (n > 150 cells). (B) Cells expressing control shRNA or shRNA targeting mH2A1 were irradiated (10Gy) and analyzed by western blot with antibodies to phosphoserine 1981 of ATM (pATM), ATM, mH2A1 and tubulin (loading control). (C) Cells expressing control shRNA or shRNA targeting mH2A1 were irradiated (2Gy), fixed and processed for immunofluorescent staining using 53BP1 antibody. The number of cells with >5 foci were counted. Results ± SD (n > 150 cells). (D) Cells expressing control shRNA or shRNA targeting mH2A1 were irradiated (10Gy) followed by western blot analysis with antibodies specific for chk2 and phospho-chk2.
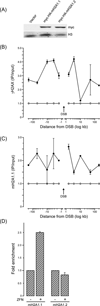
Figure 3. mH2A1.1 is recruited to DSBs
(A) Expression of myc-HA-mH2A1.1 and myc-HA-mH2A1.2 in 293T cells. (B) and (C): 293T cells expressing myc-HA-mH2A1.1 were transiently transfected with vector or p84-ZFN. 18h later cells were processed for ChIP analysis using either (B) γH2AX antibody or (C) HA antibody to detect the HA-mH2A1.1. Samples were processed for ChIP using the indicated primer pairs. ChIP data points were calculated as IP DNA/input DNA and were normalized to the uncut sample, which was assigned a value of 1. Results ± SD (n = 2). (D) 293T cells expressing myc-HA-mH2A1.1 or myc-HA-mH2A1.2 were transiently transfected with vector or p84-ZFN. 18h later cells were processed for ChIP analysis using HA antibody as described above.
Figure 4. mH2A1.1 is not incorporated into nucleosomes at DSBs
(A) 293T cells stably expressing myc-HA-mH2A1.1 were transfected with p84-ZFN as indicated. 18hr later, cells were either mock cross-linked or cross-linked with formaldehyde (X-link). Cells extracts were processed for ChIP using antibodies (Ab) to γH2AX, Ku70 or HA-mH2A1.1. Results ± SD (n = 3). (B) 293T cells expressing myc-HA-mH2A1.1 were transiently transfected with p84-ZFN in the presence or absence of PARPi (20µM AZD2281). Cells were processed for ChIP and myc-HA-mH2A1.1 accumulation at the DSB (−0.5kB) or at a distant site (-50kb) analyzed by RT-qPCR.
Similar articles
- Differential requirement for H2AX and 53BP1 in organismal development and genome maintenance in the absence of poly(ADP)ribosyl polymerase 1.
Orsburn B, Escudero B, Prakash M, Gesheva S, Liu G, Huso DL, Franco S. Orsburn B, et al. Mol Cell Biol. 2010 May;30(10):2341-52. doi: 10.1128/MCB.00091-10. Epub 2010 Mar 15. Mol Cell Biol. 2010. PMID: 20231360 Free PMC article. - Targeting BRG1 chromatin remodeler via its bromodomain for enhanced tumor cell radiosensitivity in vitro and in vivo.
Kwon SJ, Lee SK, Na J, Lee SA, Lee HS, Park JH, Chung JK, Youn H, Kwon J. Kwon SJ, et al. Mol Cancer Ther. 2015 Feb;14(2):597-607. doi: 10.1158/1535-7163.MCT-14-0372. Epub 2014 Dec 12. Mol Cancer Ther. 2015. PMID: 25504753 - Protecting the heritable genome: DNA damage response mechanisms in spermatogonial stem cells.
Rübe CE, Zhang S, Miebach N, Fricke A, Rübe C. Rübe CE, et al. DNA Repair (Amst). 2011 Feb 7;10(2):159-68. doi: 10.1016/j.dnarep.2010.10.007. Epub 2010 Nov 30. DNA Repair (Amst). 2011. PMID: 21123119 - Spatiotemporal characterization of ionizing radiation induced DNA damage foci and their relation to chromatin organization.
Costes SV, Chiolo I, Pluth JM, Barcellos-Hoff MH, Jakob B. Costes SV, et al. Mutat Res. 2010 Apr-Jun;704(1-3):78-87. doi: 10.1016/j.mrrev.2009.12.006. Epub 2010 Jan 8. Mutat Res. 2010. PMID: 20060491 Free PMC article. Review. - Chromatin dynamics in DNA double-strand break repair.
Shi L, Oberdoerffer P. Shi L, et al. Biochim Biophys Acta. 2012 Jul;1819(7):811-9. doi: 10.1016/j.bbagrm.2012.01.002. Epub 2012 Jan 17. Biochim Biophys Acta. 2012. PMID: 22285574 Free PMC article. Review.
Cited by
- Reprogramming cellular events by poly(ADP-ribose)-binding proteins.
Krietsch J, Rouleau M, Pic É, Ethier C, Dawson TM, Dawson VL, Masson JY, Poirier GG, Gagné JP. Krietsch J, et al. Mol Aspects Med. 2013 Dec;34(6):1066-87. doi: 10.1016/j.mam.2012.12.005. Epub 2012 Dec 23. Mol Aspects Med. 2013. PMID: 23268355 Free PMC article. Review. - MacroH2A1 Regulation of Poly(ADP-Ribose) Synthesis and Stability Prevents Necrosis and Promotes DNA Repair.
Ruiz PD, Hamilton GA, Park JW, Gamble MJ. Ruiz PD, et al. Mol Cell Biol. 2019 Dec 11;40(1):e00230-19. doi: 10.1128/MCB.00230-19. Print 2019 Dec 11. Mol Cell Biol. 2019. PMID: 31636161 Free PMC article. - Histone variants: the tricksters of the chromatin world.
Volle C, Dalal Y. Volle C, et al. Curr Opin Genet Dev. 2014 Apr;25:8-14,138. doi: 10.1016/j.gde.2013.11.006. Epub 2014 Jan 24. Curr Opin Genet Dev. 2014. PMID: 24463272 Free PMC article. Review. - Variants of core histones and their roles in cell fate decisions, development and cancer.
Buschbeck M, Hake SB. Buschbeck M, et al. Nat Rev Mol Cell Biol. 2017 May;18(5):299-314. doi: 10.1038/nrm.2016.166. Epub 2017 Feb 1. Nat Rev Mol Cell Biol. 2017. PMID: 28144029 Review. - Roles of Histone H2A Variants in Cancer Development, Prognosis, and Treatment.
Lai PM, Chan KM. Lai PM, et al. Int J Mol Sci. 2024 Mar 9;25(6):3144. doi: 10.3390/ijms25063144. Int J Mol Sci. 2024. PMID: 38542118 Free PMC article. Review.
References
- Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nature cell biology. 2011;13:1161–1169. - PubMed
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous