Porphyromonas gingivalis entry into gingival epithelial cells modulated by Fusobacterium nucleatum is dependent on lipid rafts - PubMed (original) (raw)
Porphyromonas gingivalis entry into gingival epithelial cells modulated by Fusobacterium nucleatum is dependent on lipid rafts
Atsushi Saito et al. Microb Pathog. 2012 Nov-Dec.
Abstract
Host cell invasion by a major periodontal pathogen, Porphyromonas gingivalis, has been proposed as an important mechanism involved in host-pathogen interactions in periodontal and cardiovascular diseases. The present study sought to gain insight into the underlying mechanism(s) involved in previously demonstrated fusobacterial modulation of host cell invasion by P. gingivalis. An immortalized human gingival cell line Ca9-22 was dually infected with P. gingivalis ATCC 33277 and Fusobacterium nucleatum TDC 100, and intracellular invasion was assessed by scanning electron microscopy (SEM) and confocal scanning laser microscopy (CSLM). SEM observation showed that P. gingivalis and F. nucleatum formed consortia and were in the process of penetrating into Ca9-22 by 30-60 min after infection. In CSLM, Ca9-22 cells that contained both P. gingivalis and F. nucleatum were frequently observed after 2 h, although cells that contained exclusively P. gingivalis were also found. Infection by P. gingivalis and/or F. nucleatum revealed evident colocalization with a lipid raft marker, GM1-containing membrane microdomains. In an antibiotic protection assay, depletion of epithelial plasma membrane cholesterol resulted in a significant reduction of recovered P. gingivalis or F. nucleatum (∼33% of untreated control; p < 0.001). This inhibition was also confirmed by CSLM. Sequential infection experiments showed that timing of infection by each species could critically influence the invasion profile. Co-infection with F. nucleatum significantly enhanced host cell invasion by P. gingivalis 33277, its serine phophatase SerB mutant and complemented strains, suggesting that the SerB does not play a major role in this fusobacterial enhancement of P. gingivalis invasion. Thus, the interaction between F. nucleatum and host cells may be important in the fusobacterial enhancement of P. gingivalis invasion. Collectively, these results suggest that lipid raft-mediated process is at least one of the potential mechanisms involved in fusobacterium-modulated host cell invasion by P. gingivalis.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
Fig. 1
(a) and (b) SEM of control uninfected Ca9-22 cells demonstrated that cellular projections (surface roughness) were not due to the presence of bacteria. ×5000 and ×10,000. (c) Ca9-22 co-infected with P. gingivalis ATCC 33277 and F. nucleatum TDC 100 for 1 h. P. gingivalis and F. nucleatum were commonly forming consortia and appear to be penetrating into the host cell. ×10,000 (d) Enlarged view; membrane blebbing was observed (arrow). (e) Near the entry site of F. nucleatum_–_P. gingivalis consortia, independent P. gingivalis (arrow) was observed.
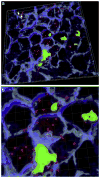
Fig. 2
(a) Invasion of P. gingivalis or F. nucleatum into gingival epithelial cells. Z stacks of the X_–_Y sections of CSLM were processed to render a 3D image using ‘Iso Surface’ functions of Imaris 7.0.0 software. Internalized P. gingivalis ATCC 33277 and F. nucleatum TDC 100 (shown as longer, fusiform shape) were stained red, while extracellular bacteria were shown green-yellow. Anti-P. gingivalis and anti-F. nucleatum antisera were used. ‘Spot Detection’ algorithm of Imaris software was used to further differentiate P. gingivalis from F. nucleatum. The host cell cytoskeleton stained with phalloidin appeared blue. (b) Enlarged view. (For interpretation of color referred in this figure legend, the reader is referred to web version of the article.)
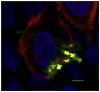
Fig. 3
P. gingivalis and F. nucleatum colocalizes with lipid rafts. Ca9-22 cells, cultured on glass coverslips, were exposed to P. gingivalis ATCC 33277 and F. nucleatum TDC 100 (MOI = 100:1). Dual species infection was allowed to proceed for 15 min and unattached bacteria were removed by washing, followed by cell fixation and staining for bacteria (anti-P. gingivalis and anti-F. nucleatum antibodies + Alexa Fluor 488- conjugated goat anti-rabbit IgG; green), and GM1 (lipid raft marker) with Alexa Fluor 594-labeled CTB (red). CSLM revealed apparent colocalization between P. gingivalis and F. nucleatum (appearing yellow) and GM1-containing membrane microdomains (red). DAPI (Blue). Bar = 5 μm.
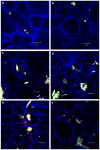
Fig. 4
Effect of cholesterol depletion on the bacterial invasion assessed by CSLM. Ca9-22 cells were incubated with DMSO (control) or 10 mM methyl-beta-cyclodextrin (MβCD) or for 30 min at 37 °C, prior to infection. (a) P. gingivalis 33277 control, (b) P. gingivalis + MβCD, (c) F. nucleatum TDC 100 control, (d) F. nucleatum TDC 100 + MβCD, (e) P. gingivalis + F. nucleatum control, (f) P. gingivalis + F. nucleatum + MβCD. Bar = 10 μm. Internalized bacteria were stained red, while extracellular bacteria were shown green-yellow. For dual-infection, anti-P. gingivalis and anti-F. nucleatum antisera were used. The host cell cytoskeleton stained with phalloidin appeared blue. (For interpretation of color referred in this figure legend, the reader is referred to web version of the article.)
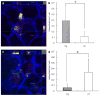
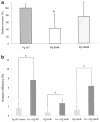
Fig. 5
Effect of F. nucleatum internalization on P. gingivalis invasion. (a) Ca9-22 cells were co-infected with F. nucleatum TDC 100 and P. gingivalis 33277 for 2 h. It was common to find both P. gingivalis and F. nucleatum intracellulary, but some cells exclusively contained P. gingivalis (data not shown). Internalized bacteria were stained red, while extracellular bacteria were shown green-yellow. The host cell cytoskeleton stained with phalloidin appeared blue. (b) Quantification of intracellular bacteria. Pg: P. gingivalis 33277, Fn: F. nucleatum TDC 100. Values are shown as means ± standard deviations. At least five fields and ~200 cells were analyzed for each experiment, and three independent experiments were done. Statistical significance between invasion levels was measured by using Mann–Whitney U test. *p < 0.05. (c) Ca9-22 cells were first infected with F. nucleatum TDC 100 for 1 h, then P. gingivalis 33277 was added and further incubated for another hour. In this sequential infection experiment, intracellular bacteria were predominantly F. nucleatum. Internalized bacteria were stained red, while extracellular bacteria were shown green-yellow. Anti-P. gingivalis and anti-F. nucleatum antisera were used. The host cell cytoskeleton stained with phalloidin appeared blue. (d) Quantification of intracellular bacteria. Values are shown as means ± standard deviations. *p < 0.05. (For interpretation of color referred in this figure legend, the reader is referred to web version of the article.)
Fig. 6
(a) Invasion of gingival epithelial cells (Ca9-22) by P. gingivalis 33277 wild type strain, its isogenic SerB mutant and complemented strains. Statistical significance between invasion levels was measured by using Mann–Whitney U test. *p < 0.05. Pg WT: P. gingivalis 33277, Pg SerB-: P. gingivalis SerB mutant, Pg cSerB: Complemented SerB mutant. (b) Effect of co-infection with F. nucleatum on invasion of gingival epithelial cells by P. gingivalis 33277, its isogenic mutant and complemented strains. Statistical significance between invasion levels was measured by using Kruskal–Wallis test with Dunn’s multiple comparisons test. *p < 0.001. Fn: F. nucleatum TDC 100.
Similar articles
- Fusobacterium nucleatum enhances invasion of human gingival epithelial and aortic endothelial cells by Porphyromonas gingivalis.
Saito A, Inagaki S, Kimizuka R, Okuda K, Hosaka Y, Nakagawa T, Ishihara K. Saito A, et al. FEMS Immunol Med Microbiol. 2008 Dec;54(3):349-55. doi: 10.1111/j.1574-695X.2008.00481.x. FEMS Immunol Med Microbiol. 2008. PMID: 19049647 - Differential ability of periodontopathic bacteria to modulate invasion of human gingival epithelial cells by Porphyromonas gingivalis.
Saito A, Inagaki S, Ishihara K. Saito A, et al. Microb Pathog. 2009 Dec;47(6):329-33. doi: 10.1016/j.micpath.2009.09.012. Epub 2009 Oct 7. Microb Pathog. 2009. PMID: 19818393 - Relatively low invasive capacity of Porphyromonas gingivalis strains into human gingival fibroblasts in vitro.
Jang JY, Baek KJ, Choi Y, Ji S. Jang JY, et al. Arch Oral Biol. 2017 Nov;83:265-271. doi: 10.1016/j.archoralbio.2017.08.007. Epub 2017 Aug 16. Arch Oral Biol. 2017. PMID: 28841474 - Immunological Pathways Triggered by Porphyromonas gingivalis and Fusobacterium nucleatum: Therapeutic Possibilities?
de Andrade KQ, Almeida-da-Silva CLC, Coutinho-Silva R. de Andrade KQ, et al. Mediators Inflamm. 2019 Jun 24;2019:7241312. doi: 10.1155/2019/7241312. eCollection 2019. Mediators Inflamm. 2019. PMID: 31341421 Free PMC article. Review. - About a Possible Impact of Endodontic Infections by Fusobacterium nucleatum or Porphyromonas gingivalis on Oral Carcinogenesis: A Literature Overview.
Ciani L, Libonati A, Dri M, Pomella S, Campanella V, Barillari G. Ciani L, et al. Int J Mol Sci. 2024 May 7;25(10):5083. doi: 10.3390/ijms25105083. Int J Mol Sci. 2024. PMID: 38791123 Free PMC article. Review.
Cited by
- An in vitro model of Fusobacterium nucleatum and Porphyromonas gingivalis in single- and dual-species biofilms.
Tavares LJ, Klein MI, Panariello BHD, Dorigatti de Avila E, Pavarina AC. Tavares LJ, et al. J Periodontal Implant Sci. 2018 Feb 27;48(1):12-21. doi: 10.5051/jpis.2018.48.1.12. eCollection 2018 Feb. J Periodontal Implant Sci. 2018. PMID: 29535887 Free PMC article. - Insights into the oral microbiota in human systemic cancers.
Su L, Yang R, Sheng Y, Ullah S, Zhao Y, Shunjiayi H, Zhao Z, Wang Q. Su L, et al. Front Microbiol. 2024 May 2;15:1369834. doi: 10.3389/fmicb.2024.1369834. eCollection 2024. Front Microbiol. 2024. PMID: 38756728 Free PMC article. Review. - Differential involvement of the canonical and noncanonical inflammasomes in the immune response against infection by the periodontal bacteria Porphyromonas gingivalis and Fusobacterium nucleatum.
De Andrade KQ, Almeida-da-Silva CLC, Ojcius DM, Coutinho-Silva R. De Andrade KQ, et al. Curr Res Microb Sci. 2021 Feb 23;2:100023. doi: 10.1016/j.crmicr.2021.100023. eCollection 2021 Dec. Curr Res Microb Sci. 2021. PMID: 34841314 Free PMC article. - Polymicrobial synergy and dysbiosis in inflammatory disease.
Lamont RJ, Hajishengallis G. Lamont RJ, et al. Trends Mol Med. 2015 Mar;21(3):172-83. doi: 10.1016/j.molmed.2014.11.004. Epub 2014 Nov 20. Trends Mol Med. 2015. PMID: 25498392 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources