Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation - PubMed (original) (raw)
Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation
Jeff R Broadbent et al. BMC Genomics. 2012.
Abstract
Background: The broad ecological distribution of L. casei makes it an insightful subject for research on genome evolution and lifestyle adaptation. To explore evolutionary mechanisms that determine genomic diversity of L. casei, we performed comparative analysis of 17 L. casei genomes representing strains collected from dairy, plant, and human sources.
Results: Differences in L. casei genome inventory revealed an open pan-genome comprised of 1,715 core and 4,220 accessory genes. Extrapolation of pan-genome data indicates L. casei has a supragenome approximately 3.2 times larger than the average genome of individual strains. Evidence suggests horizontal gene transfer from other bacterial species, particularly lactobacilli, has been important in adaptation of L. casei to new habitats and lifestyles, but evolution of dairy niche specialists also appears to involve gene decay.
Conclusions: Genome diversity in L. casei has evolved through gene acquisition and decay. Acquisition of foreign genomic islands likely confers a fitness benefit in specific habitats, notably plant-associated niches. Loss of unnecessary ancestral traits in strains collected from bacterial-ripened cheeses supports the hypothesis that gene decay contributes to enhanced fitness in that niche. This study gives the first evidence for a L. casei supragenome and provides valuable insights into mechanisms for genome evolution and lifestyle adaptation of this ecologically flexible and industrially important lactic acid bacterium. Additionally, our data confirm the Distributed Genome Hypothesis extends to non-pathogenic, ecologically flexible species like L. casei.
Figures
Figure 1
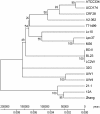
Alignment of L . casei genomes. Five closed and 12 new draft genomes were aligned using progressive Mauve. Three boxed areas outline large highly collinear regions where the order of Locally Collinear Blocks is highly conserved among all 17 strains. The boxes are a fixed length, so that all region size can be compared, and maps were adjusted to the left to reflect length differences. Vertical red lines indicate contig boundaries, and unplaced contigs were sorted to the right end of each map.
Figure 2
Hierarchical clustering of 17 L . casei strains based on overall gene content. Members of each cluster were aligned using Mauve, and vertical red lines indicate contig boundaries. Locally Collinear Blocks (LCBs) are colored to reveal harmonization within each cluster, but do not have identity to LCBs of the same color outside a particular cluster. The Lactobacillus casei BL23 genome was used as a reference to order and orientate contigs for strains included in all clusters except B, where strains 21/1, 32G, and Lc-10 were ordered using the L. casei Zhang genome as a reference, and strains CRF28 and 12A were ordered based on L. casei ATCC 334. The BL23 alignment is shown with cluster B to compare genomic similarity.
Figure 3
Genetic relationships among 17 L . casei strains as defined by multilocus sequence typing. Concatenated sequences of 6 MLST loci (ftsZ, metRS, mutL, pgm, nrdD, and polA) were analyzed as described previously [30].
Figure 4
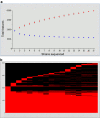
Feature numbers for the core and pan-genome across 17 L . casei strains. Panel A shows numbers of total features in the core (blue) and pan-(red) genome as a function of the number of strains sequenced. The average of 500 random permutations of the genome order is presented for the pan and core genome content; the error bars represent the standard deviation of these results. The heat map in panel B illustrates the distribution of core and accessory genes (combined in red) across the 17 sequenced strains. New accessory genes that are contributed to the pan-genome by each strain's sequence are depicted as a red cap at the top of each strain name. Black regions under that cap indicate features missing in that strain but present in one or more of the other sequenced L. casei.
Figure 5
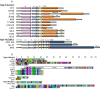
Graphic representation of CRISPR elements in L . casei genomes. Panel A, graphic representation of CRISPR-cas loci. Top, Type II-A CRISPR-Cas systems; Bottom, Type I-E CRISPR-Cas systems. cas genes are represented by colored arrows, while CRISPR repeat-spacer arrays are represented as orange (top) and blue (bottom) boxes. IS elements are colored in green. Numbered genes represented by narrow box arrows refer to previously published nomenclature. Homologous genes are represented using identical colors. Hashed elements represent gaps in the genome draft sequence. Panel B, graphic representation of CRISPR spacers across the two CRISPR-Cas types found in L. casei. Each unique spacer sequence is represented as a specific combination of two colors. Repeats are not included. Missing spacers are represented by crossed squares.
Figure 6
CRISPR spacer homology to genetic elements and proto-spacer associated motifs. Left, top 10 matches between CRISPR spacers and corresponding proto-spacers in phages, plasmids and chromosomal sequences. Numbers indicate the spacer position from the leader end. Levels of sequence similarity, location of the start and end of the match, and sequence immediately upstream (START-7) or immediately downstream (END+7) are provided. Right, conservation of certain nucleotides in the immediate vicinity of the proto-spacer sequence.
Figure 7
L . casei gene clusters that may function in cellobiose uptake and hydrolysis. Asterisks identify clusters that are predicted to be nonfunctional in particular strains due to the presence of one or more pseudogenes. Abbreviations: P-β-Glu, phospho-β-glucosidase; β-Glu, β-glucosidase; ABC-ATP, ABC-P, and ABC-SB represent ATP-binding, periplasmic, and substrate-binding components, respectively, of an ABC transport system; EIIABC, EIIA, EIIB, EIIBC, or EIIC, represent sugar-specific enzymes for a PTS transport system; AT, antiterminator protein; TR, transcriptional regulator; Mem Prt, predicted membrane protein of unknown function; Hyp Prt, hypothetical protein; N-AcM K, N-acetymannosamine kinase. Locus tags in L. casei ATCC 334 for the glucosidase CDS in clusters 1–5 are LSEI_0448, LSEI_0700, LSEI_1104, LSEI_1778, and LSEI_2191, respectively. Locus tags in 21/1 for the glucosidase CDS in clusters 6–8 are LCA211_0376, LCA211_0004, and LCA211_2196, respectively. The locus tag in T71499 for the glucosidase CDS in cluster 9 is LCAT71499_1435.
Similar articles
- Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution.
Cai H, Thompson R, Budinich MF, Broadbent JR, Steele JL. Cai H, et al. Genome Biol Evol. 2009 Jul 14;1:239-57. doi: 10.1093/gbe/evp019. Genome Biol Evol. 2009. PMID: 20333194 Free PMC article. - Genomic adaptation of the Lactobacillus casei group.
Toh H, Oshima K, Nakano A, Takahata M, Murakami M, Takaki T, Nishiyama H, Igimi S, Hattori M, Morita H. Toh H, et al. PLoS One. 2013 Oct 8;8(10):e75073. doi: 10.1371/journal.pone.0075073. eCollection 2013. PLoS One. 2013. PMID: 24116025 Free PMC article. - Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity.
Cai H, Rodríguez BT, Zhang W, Broadbent JR, Steele JL. Cai H, et al. Microbiology (Reading). 2007 Aug;153(Pt 8):2655-2665. doi: 10.1099/mic.0.2007/006452-0. Microbiology (Reading). 2007. PMID: 17660430 - Multilocus genotyping for classification and genetic structuring of Lactobacillus casei: insights from source and geographical origin.
Tynybayeva I, Tekebayeva Z, Bissenova G, Mynbayeva M, Sarmurzina Z. Tynybayeva I, et al. World J Microbiol Biotechnol. 2024 Feb 13;40(3):93. doi: 10.1007/s11274-024-03898-z. World J Microbiol Biotechnol. 2024. PMID: 38349588 Review. - Beyond the pan-genome: current perspectives on the functional and practical outcomes of the distributed genome hypothesis.
Hammond JA, Gordon EA, Socarras KM, Chang Mell J, Ehrlich GD. Hammond JA, et al. Biochem Soc Trans. 2020 Dec 18;48(6):2437-2455. doi: 10.1042/BST20190713. Biochem Soc Trans. 2020. PMID: 33245329 Free PMC article. Review.
Cited by
- Draft Genome Sequence of the Respiration-Competent Strain Lactobacillus casei N87.
Zotta T, Ricciardi A, Parente E, Reale A, Ianniello RG, Bassi D. Zotta T, et al. Genome Announc. 2016 May 5;4(3):e00348-16. doi: 10.1128/genomeA.00348-16. Genome Announc. 2016. PMID: 27151805 Free PMC article. - Extraction and Characterization of Cocoa Bean Shell Cell Wall Polysaccharides.
Younes A, Karboune S, Liu L, Andreani ES, Dahman S. Younes A, et al. Polymers (Basel). 2023 Feb 1;15(3):745. doi: 10.3390/polym15030745. Polymers (Basel). 2023. PMID: 36772046 Free PMC article. - Conversion of Methionine to Cysteine in Lactobacillus paracasei Depends on the Highly Mobile cysK-ctl-cysE Gene Cluster.
Wüthrich D, Irmler S, Berthoud H, Guggenbühl B, Eugster E, Bruggmann R. Wüthrich D, et al. Front Microbiol. 2018 Oct 17;9:2415. doi: 10.3389/fmicb.2018.02415. eCollection 2018. Front Microbiol. 2018. PMID: 30386310 Free PMC article. - Population genomics of Lacticaseibacillus paracasei: pan-genome, integrated prophage, antibiotic resistance, and carbohydrate utilization.
Pei Z, Li X, Cui S, Yang B, Lu W, Zhao J, Mao B, Chen W. Pei Z, et al. World J Microbiol Biotechnol. 2023 Aug 17;39(10):280. doi: 10.1007/s11274-023-03722-0. World J Microbiol Biotechnol. 2023. PMID: 37587248 - Comparative genomic and metabolic analysis of three Lactobacillus paracasei cheese isolates reveals considerable genomic differences in strains from the same niche.
Stefanovic E, McAuliffe O. Stefanovic E, et al. BMC Genomics. 2018 Mar 20;19(1):205. doi: 10.1186/s12864-018-4586-0. BMC Genomics. 2018. PMID: 29554864 Free PMC article.
References
- Axelsson L. In: Lactic acid bacteria microbiological and functional aspects. 3. Salminen S, Wright A, Ouwehand A, editor. Marcel Dekker Inc, New York; 2004. Lactic acid bacteria: classification and physiology; pp. 1–66.
- Kandler O, Weiss N. In: Bergey’s manual of systematic bacteriology. 9. Sneath PHA, Mair NS, Sharpe ME, Holt JG, editor. Williams and Wilkins, Baltimore; 1986. Genus Lactobacillus; pp. 1063–1065.
- Beresford T, Williams A. In: Cheese: Chemistry, Physics and Microbiology. 3. Fox PF, McSweeney PLH, Cogan TM, Guinee TP, editor. Elsevier, Amsterdam, the Netherlands; 2004. The microbiology of cheese ripening; pp. 287–318.
- Judicial Commission of the International Committee on Systematics of Bacteria. The type strain of Lactobacillus casei is ATCC 393, ATCC 334 cannot serve as the type because it represents a different taxon, the name Lactobacillus paracasei and its subspecies names are not rejected and the revival of the name ‘Lactobacillus zeae’ contravenes Rules 51b (1) and (2) of the International Code of Nomenclature of Bacteria. Opinion 82. Int J Syst Evol Microbiol. 2008;58:1764–1765. - PubMed
- Goldin BR, Gorbach SL. In: Probiotics: the scientific basis. Fuller R, editor. Chapman and Hall, London; 1992. Probiotics for humans; pp. 355–376.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases