Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells - PubMed (original) (raw)
Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells
Nicola Festuccia et al. Cell Stem Cell. 2012.
Abstract
Embryonic stem cell (ESC) self-renewal efficiency is determined by the level of Nanog expression. However, the mechanisms by which Nanog functions remain unclear, and in particular, direct Nanog target genes are uncharacterized. Here we investigate ESCs expressing different Nanog levels and Nanog(-/-) cells with distinct functionally inducible Nanog proteins to identify Nanog-responsive genes. Surprisingly, these constitute a minor fraction of genes that Nanog binds. Prominent among Nanog-reponsive genes is Estrogen-related receptor b (Esrrb). Nanog binds directly to Esrrb, enhances binding of RNAPolII, and stimulates Esrrb transcription. Overexpression of Esrrb in ESCs maintains cytokine-independent self-renewal and pluripotency. Remarkably, this activity is retained in Nanog(-/-) ESCs. Moreover, Esrrb can reprogram Nanog(-/-) EpiSCs and can rescue stalled reprogramming in Nanog(-/-) pre-iPSCs. Finally, Esrrb deletion abolishes the defining ability of Nanog to confer LIF-independent ESC self-renewal. These findings are consistent with the functional placement of Esrrb downstream of Nanog.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
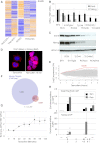
Identification of Nanog Target Genes Including Esrrb (A) Deep-SAGE profile of sorted Nanog-positive (GFP+) and Nanog-negative (GFP–) TNG cells, ESCs with wild-type levels of Nanog expression (RCN(t)) and _Nanog_−/− ESCs (RCNβH(t)). Genes were ranked according to the expression level and fold difference in expression in TNG+ versus TNG− and RCN(t) versus RCNβH(t); the plot shows the first 250 most upregulated (top) or downregulated (bottom) genes. Colors: yellow, expression above average; blue, below average. (B) Esrrb transcript levels in two cell lines overexpressing Nanog (EF4 and RCN), two cell lines with wild-type Nanog (E14Tg2a and RCN(t)), two Nanog+/− cell lines (TβC44 and RCNβ(t)), and two _Nanog_−/− cell lines (TβC44Cre6 and RCNβH(t)). Error bars: standard deviation (n = 4). (C) Immunoblot analysis of Esrrb and Nanog levels in the same ESC lines. (D) Immunohistochemical analysis of the intracellular localization of Nanog in ESΔN-NERT cells in response to 1 μM tamoxifen as indicated. (E) Global transcriptional changes after ESΔN-NERT stimulation with tamoxifen as indicated; the Esrrb changes are in red. Mean expression levels in three independent experiments are shown. (F) Venn diagram showing the intersection of significantly upregulated or downregulated genes identified in (E) compared to genes bound by Nanog according to two independent genome-wide ChIP studies. (G) Esrrb pre-mRNA kinetics in ESΔN-NERT cells stimulated with tamoxifen as indicated. Error bars: standard deviation of expression values in three different clones. (H) Chromatin from ESΔN-NERT cells treated with 1 μM tamoxifen for 0 or 24 hr was immunoprecipitated with Nanog or total RNAPolII antibodies. Enrichment relative to the ArpP0 promoter is measured using the primers indicated at Esrrb. Error bars: standard deviation (n = 3); ∗p ≤ 0.05, ∗∗p ≤ 0.01. See also Figures S1 and S2 and Tables S1.1 and 1.2.
Figure 2
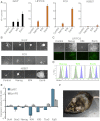
Esrrb Overexpression Confers LIF and Nanog-Independent Self-Renewal (A) _lifr_−/−:PyLT+ LRK1 cells were transfected with episomal plasmids encoding the indicated ORF (EV; empty vector) and the number of AP-positive colonies was determined after clonal density plating in the absence of IL-6/sIL6R. Error bars: standard deviation (n = 3). (B) Schematic representation of EfEsrrb ESCs. (C) Colony morphology (top) and AP staining (bottom) of EfEsrrb c1 cultured in the presence of hLIF-05. (D) E14Tg2a, Nanog-, and Esrrb-overexpressing cells before and after Cre reversion were plated at clonal density and cultured in the presence or absence of LIF or hLIF-05 for 7 days, and the number of AP-positive colonies was counted. Error bars: standard deviation (n = 3). (E) Chimeras generated after injection into C57BL/6 blastocysts of EfEsrrb-Cre ESCs passaged twice at clonal density in the presence of hLIF-05 and transfected with a Cre expression vector to excise the Esrrb transgene. (F) Schematic representation of the genetic manipulations used to make ESΔN-iNanog or ESΔN-iEsrrb cells. (G) Colony morphology of ESΔN-iNanog (iN) or ESΔN-iEsrrb (iE) cells plated at clonal density and cultured in the presence of hLIF-05 (+/− doxycycline) for 8 days. Right hand panels: AP staining of colonies formed in the presence of doxycycline. (H) Number of AP-positive colonies formed after clonal density plating of ESΔN-iNanog (iN) or ESΔN-iEsrrb (iE) cells in the presence of LIF or hLIF-05 and cultured (+/− doxycycline) for 8 days. Error bars: standard deviation (n = 3). (I) ESΔN-iNanog (iN) and ESΔN-iEsrrb (iE) cells in a neural differentiation protocol, without (top rows) or with (bottom rows) doxycycline for 9 days. Cells were fixed, stained for βIII-Tubulin (Tuj), and analyzed by fluorescence microscopy. See also Figures S3 and S4 and Table S2.
Figure 3
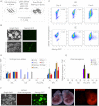
Expression of Esrrb Reverts EpiSCs to Chimera Competency (A) AP-positive colony formation by Epi-iPSCs. EpiSCs expressing polyoma large T-antigen were transfected with episomal vectors encoding empty vector (EV), Nanog, Klf4, or Esrrb, plated in the indicated medium containing puromycin, and stained for AP after 7 days. Error bars: standard deviation (n = 3). (B) Morphology of primary Epi-iPSC colonies formed after transfection of the respective episomal vector and culture in the indicated medium for 7 days. (C) Morphology and Nanog:GFP expression of primary Epi-iPSC colonies formed after transfection of the respective episomal vector and culture in FCS/LIF/GMEMβ for 7 days. (D) FACS analysis of Pecam1 expression 7 days after transfection of the indicated DNAs. TNG/T ESCs (blue) and EpiSCs (gray) were used as controls for Pecam1 expression. (E) mRNA expression in E14/T EpiSC and Epi-iPSC colonies expanded in the absence of selection after episomal expression of Esrrb and medium switch into FCS/LIF/GMEMβ. Error bars: standard deviation of gene expression in three independent experiments. (F) Chimeric mouse obtained from blastocyst injection of Esrrb-induced Epi-iPSCs. See also Figure S5 and Table S3.
Figure 4
Nanog Null EpiSC Are Reverted to Naive Pluripotency by Esrrb Expression (A) _Nanog_−/− EpiSCs carrying doxycycline-inducible Nanog or Esrrb transgenes were plated in FCS/LIF/GMEMβ with doxycycline for the indicated times. After 7 days, plates were stained for AP. (B) Scoring of the AP colonies obtained from the experiment described in (A). Error bars: standard deviation (n = 3). (C) mRNA expression in uninduced EpiΔN-iEsrrb and the reverted Epi-iPSΔN-iEsrrb ESC-like colonies obtained by induction of Esrrb and expansion in the absence of selection and doxycycline. Error bars: standard deviation of gene expression in two independent experiments. (D) Brightfield (top panels) and fluorescence (bottom panels) images of ESΔN-iEsrrb, EpiΔN-iEsrrb, and Epi-iPSΔN-iEsrrb cells. (E) AP-positive colonies of Epi-iPSΔN-iEsrrb cells grown in N2B27 supplemented with BMP/LIF (top) or 2i/LIF (bottom). (F) Chimeric mouse obtained from a blastocyst injection with Epi-iPSΔN-iEsrrb cells. See also Table S3.
Figure 5
In Vitro Reprogramming by Cell Fusion Can Proceed in the Absence of Nanog (A) Schematic representation of the genetic manipulations performed on the lines used in the fusion experiments: ESΔN-iNanog and ESΔN-Esrrb cells and RCNβH(t) Red NSCs. (B) Colonies formed by ESΔN-iNanog (iN) or ESΔN-iEsrrb (iE) × RCNβH(t) Red NSCs hybrids after 16 days selection in blasticidin/hygromycin in the presence or absence of doxycycline. (C) Morphology of ESΔN-iEsrrb (iE) × RCNβH(t) Red NSC hybrids cultured in doxycycline or released from doxycycline for three passages (10 days) in the presence or absence of G418 to select for active Nanog transcription. (D) Gene expression profiles of endogenous genes in RCNβH(t) Red NSCs, ESΔN-iNanog (iN) cells or ESΔN-iEsrrb (iE) cells, and hybrid lines after three passages in the indicated conditions. Primers do not detect transgenes. Nanog primers bind to intron I, which is not deleted in the targeted alleles. Transcript levels are normalized to TBP and relative to expression in RCNβH(t) Red NS (Olig2) or ESΔN-iNanog cells cultured in G418 (all other genes). Error bars: ESC × NSC hybrids: standard deviation of gene expression in three independent clones. ESC and NSC lines: standard deviation of gene expression in two independent experiments. See also Figure S6 and Tables S4, S5, and S6.
Figure 6
Esrrb Can Reprogram _Nanog_−/− Somatic Cells to Naive Pluripotency (A) Experimental scheme used to derive pre-iPSCs and to induce completion of reprogramming. (B) Morphology and Nanog:GFP expression in pre-iPSΔN-iEsrrb cells cultured in the absence of doxycycline (top) or in doxycycline/5-azacytidine for 3 days (bottom). (C) FACS plots of viral transgene expression (dsRed) and Nanog:GFP in pre-iPSΔN-iNanog (iN) or pre-iPSΔN-iEsrrb (iE) cells treated with doxycycline/5-azacytidine as indicated. Percentages of cells positive for Nanog:GFP are shown. (D) Q-PCR of endogenous genes in ESΔN-iNanog (iN) or ESΔN-iEsrrb (iE) cells and derivative NSCs, pre-iPSCs, and iPSCs. Primers do not detect transgenes. Nanog primers bind to intron I, which remains in all targeted cells. All cell lines were maintained without doxycycline for at least three passages. mRNA levels (normalized to TBP) are relative to expression in NSΔN-iEsrrb cells (Olig2) or ESΔN-iEsrrb cells (all other genes). Error bars: iPSCs: standard deviation of gene expression in three independent clones. ESC, pre-iPSC, and NSC lines: standard deviation of gene expression in three independent experiments. (E) Q-PCR of retroviral transgenes in ESΔN-iNanog (iN) or ESΔN-iEsrrb (iE) cells and derivative NSCs, pre-iPSCs, and iPSCs. Primers do not detect endogenous transcripts. mRNA levels (normalized to TBP) are relative to expression in pre-iPSΔN-iEsrrb cells. Error bars: standard deviation of expression values in three independent experiments. (F) Morphology, dsRed, and Nanog:GFP expression in iPSΔN-iEsrrb cells cultured on gelatin without doxycycline for three passages. (G) Midgestation embryo obtained from blastocyst injection of iPSΔN-iEsrrb cells transfected with a ubiquitously expressed TdTomato transgene (right); control embryo (left). See also Table S7.
Figure 7
Loss of Esrrb Impairs Nanog-Driven LIF Independence (A) Schematic representation of the genetic manipulations used to make conditional knockout (_Esrrb_f/fn) ESCs that have two floxed Esrrb alleles and express Cre-ERT2. (B) Morphology and expression of Oct4 and Esrrb in _Esrrb_f/fn and deleted _Esrrb_Δ/Δ lines. (C) Colony formation after clonal density plating and 7 days culture (+/− LIF; values are the average of six independent clones for each indicated line). Error bars: standard deviation of the results obtained from six clones each analyzed in triplicate. (D) Representative morphologies of colonies formed by the indicated lines after 7 days of culture (+/− LIF). See also Figure S7.
Comment in
- Pluripotency network in embryonic stem cells: maybe Leibniz was right all along.
Zwaka TP. Zwaka TP. Cell Stem Cell. 2012 Oct 5;11(4):441-2. doi: 10.1016/j.stem.2012.09.005. Cell Stem Cell. 2012. PMID: 23040470
Similar articles
- Esrrb extinction triggers dismantling of naïve pluripotency and marks commitment to differentiation.
Festuccia N, Halbritter F, Corsinotti A, Gagliardi A, Colby D, Tomlinson SR, Chambers I. Festuccia N, et al. EMBO J. 2018 Nov 2;37(21):e95476. doi: 10.15252/embj.201695476. Epub 2018 Oct 1. EMBO J. 2018. PMID: 30275266 Free PMC article. - Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming.
Percharde M, Lavial F, Ng JH, Kumar V, Tomaz RA, Martin N, Yeo JC, Gil J, Prabhakar S, Ng HH, Parker MG, Azuara V. Percharde M, et al. Genes Dev. 2012 Oct 15;26(20):2286-98. doi: 10.1101/gad.195545.112. Epub 2012 Sep 26. Genes Dev. 2012. PMID: 23019124 Free PMC article. - Esrrb activates Oct4 transcription and sustains self-renewal and pluripotency in embryonic stem cells.
Zhang X, Zhang J, Wang T, Esteban MA, Pei D. Zhang X, et al. J Biol Chem. 2008 Dec 19;283(51):35825-33. doi: 10.1074/jbc.M803481200. Epub 2008 Oct 28. J Biol Chem. 2008. PMID: 18957414 - The genetics of induced pluripotency.
Ralston A, Rossant J. Ralston A, et al. Reproduction. 2010 Jan;139(1):35-44. doi: 10.1530/REP-09-0024. Reproduction. 2010. PMID: 19605512 Review. - Nanog and transcriptional networks in embryonic stem cell pluripotency.
Pan G, Thomson JA. Pan G, et al. Cell Res. 2007 Jan;17(1):42-9. doi: 10.1038/sj.cr.7310125. Cell Res. 2007. PMID: 17211451 Review.
Cited by
- Molecular network of miR-1343 regulates the pluripotency of porcine pluripotent stem cells via repressing OTX2 expression.
Xie Y, Cao H, Zhang Z, Zhang S, Wang H. Xie Y, et al. RNA Biol. 2019 Jan;16(1):82-92. doi: 10.1080/15476286.2018.1559688. Epub 2018 Dec 27. RNA Biol. 2019. PMID: 30567463 Free PMC article. - Epigenetic Regulation of Driver Genes in Testicular Tumorigenesis.
von Eyben FE, Kristiansen K, Kapp DS, Hu R, Preda O, Nogales FF. von Eyben FE, et al. Int J Mol Sci. 2023 Feb 19;24(4):4148. doi: 10.3390/ijms24044148. Int J Mol Sci. 2023. PMID: 36835562 Free PMC article. Review. - Targeting cancer stem cells: emerging role of Nanog transcription factor.
Wang ML, Chiou SH, Wu CW. Wang ML, et al. Onco Targets Ther. 2013 Sep 4;6:1207-20. doi: 10.2147/OTT.S38114. Onco Targets Ther. 2013. PMID: 24043946 Free PMC article. Review. - Dax1 and Nanog act in parallel to stabilize mouse embryonic stem cells and induced pluripotency.
Zhang J, Liu G, Ruan Y, Wang J, Zhao K, Wan Y, Liu B, Zheng H, Peng T, Wu W, He P, Hu FQ, Jian R. Zhang J, et al. Nat Commun. 2014 Oct 6;5:5042. doi: 10.1038/ncomms6042. Nat Commun. 2014. PMID: 25284313 Free PMC article. - Molecular control of induced pluripotency.
Theunissen TW, Jaenisch R. Theunissen TW, et al. Cell Stem Cell. 2014 Jun 5;14(6):720-34. doi: 10.1016/j.stem.2014.05.002. Cell Stem Cell. 2014. PMID: 24905163 Free PMC article. Review.
References
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. - PubMed
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials