Heparan sulfate biosynthesis: regulation and variability - PubMed (original) (raw)
Review
Heparan sulfate biosynthesis: regulation and variability
Johan Kreuger et al. J Histochem Cytochem. 2012 Dec.
Abstract
Nearly all vertebrate cells have been shown to express heparan sulfate proteoglycans (HSPGs) at the cell surface. The HSPGs bind to many secreted signaling proteins, including numerous growth factors, cytokines, and morphogens, to affect their tissue distribution and signaling. The heparan sulfate (HS) chains may have variable length and may differ with regard to both degree and pattern of sulfation. As the sulfation pattern of HS chains in most cases will determine if an interaction with a potential ligand will take place, as well as the affinity of the interaction, a key to understanding the function of HSPGs is to clarify how HS biosynthesis is regulated in different biological contexts. This review provides an introduction to the current understanding of HS biosynthesis and its regulation, and identifies research areas where more knowledge is needed to better understand how the HS biosynthetic machinery works.
Conflict of interest statement
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures
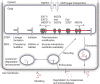
Figure 1.
Heparan sulfate (HS) structure and biosynthesis scheme. Shown is a simplified scheme outlining the different steps of HS biosynthesis involving specific enzymes or enzyme families. The structure of HS is variable, and a hypothetical example is shown. The saccharide units corresponding to symbols used are defined below the scheme. The abbreviations related to structure are as follows: NS, _N_-sulfated GlcN; 6S, 6-_O_-sulfated GlcN; 2S, 2-_O_-sulfated IdoA; 3S, 3-_O_-sulfated GlcN; Ser, serine. For additional information, see Figure 2 and the main text.
Figure 2.
Formation and fate of heparan sulfate (HS). The formation of HS takes place in the Golgi network, where most of the biosynthetic enzymes are anchored to the Golgi membrane. Biosynthetic precursors (3′-phosphoadenosine-5′-phosphosulfate [PAPS] and UDP-sugars) are formed in the cytosol and transported into the Golgi. Prior to HS polymerization, the linkage region is formed attached to a serine residue in a core protein. Next, the EXT1/EXT2 polymerase complex adds alternating units of GlcNAc and GlcA to the non-reducing end of the growing chain (arrow a indicates the direction of polymerization). The polymerization is followed by a series of modification reactions, likely to begin with_N_-deacetylation/_N_-sulfation, followed by epimerization and 2-_O_-sulfation, and finally 6-_O_- and 3-_O_-sulfation. Notably, it has recently been proposed that the direction of _N_-deacetylation/_N_-sulfation is opposite to that of polymerization (arrow b). Known interactions between enzymes are indicated, but additional protein interactions as well as larger GAGosome complexes encompassing many enzymes may exist. After completion of the modification process, the core proteins are transported to the cell membrane, where they are exocytosed. HS chains of both membrane-intercalated and secreted proteoglycans (PGs) can be trimmed by the actions of heparanase and endosulfatases, and surface-bound PGs can also be shed. Finally, endocytosis of PGs leads to degradation of HS by exoenzymes in lysosomes or, alternatively, to recycling and possibly additional rounds of HS biosynthesis/modification onto recycled core proteins. Some regulatory steps (Reg.) during the biosynthetic process are indicated.
Similar articles
- [Heparan sulfate biosynthesis in swine arterial wall: glycosylation and sulfation].
Levy P, Picard J, Bruel A. Levy P, et al. Paroi Arterielle. 1981;7(3):113-9. Paroi Arterielle. 1981. PMID: 6461835 French. - PAPST1 regulates sulfation of heparan sulfate proteoglycans in epithelial MDCK II cells.
Dick G, Akslen-Hoel LK, Grøndahl F, Kjos I, Maccarana M, Prydz K. Dick G, et al. Glycobiology. 2015 Jan;25(1):30-41. doi: 10.1093/glycob/cwu084. Epub 2014 Aug 18. Glycobiology. 2015. PMID: 25138304 - Heparan Sulfate: Biosynthesis, Structure, and Function.
Li JP, Kusche-Gullberg M. Li JP, et al. Int Rev Cell Mol Biol. 2016;325:215-73. doi: 10.1016/bs.ircmb.2016.02.009. Epub 2016 Apr 13. Int Rev Cell Mol Biol. 2016. PMID: 27241222 Review. - "Coding" and "Decoding": hypothesis for the regulatory mechanism involved in heparan sulfate biosynthesis.
Zhang X, Wang F, Sheng J. Zhang X, et al. Carbohydr Res. 2016 Jun 16;428:1-7. doi: 10.1016/j.carres.2016.04.002. Epub 2016 Apr 8. Carbohydr Res. 2016. PMID: 27088396 Review.
Cited by
- Alterations in heparan sulfate proteoglycan synthesis and sulfation and the impact on vascular endothelial function.
Pretorius D, Richter RP, Anand T, Cardenas JC, Richter JR. Pretorius D, et al. Matrix Biol Plus. 2022 Sep 7;16:100121. doi: 10.1016/j.mbplus.2022.100121. eCollection 2022 Dec. Matrix Biol Plus. 2022. PMID: 36160687 Free PMC article. Review. - Chemistry and Function of Glycosaminoglycans in the Nervous System.
Schwartz NB, Domowicz MS. Schwartz NB, et al. Adv Neurobiol. 2023;29:117-162. doi: 10.1007/978-3-031-12390-0_5. Adv Neurobiol. 2023. PMID: 36255674 Review. - The role of innate immunity in mucopolysaccharide diseases.
Parker H, Bigger BW. Parker H, et al. J Neurochem. 2019 Mar;148(5):639-651. doi: 10.1111/jnc.14632. Epub 2018 Dec 13. J Neurochem. 2019. PMID: 30451296 Free PMC article. Review. - Heparan Sulfate as a Therapeutic Target in Tauopathies: Insights From Zebrafish.
Alavi Naini SM, Soussi-Yanicostas N. Alavi Naini SM, et al. Front Cell Dev Biol. 2018 Dec 20;6:163. doi: 10.3389/fcell.2018.00163. eCollection 2018. Front Cell Dev Biol. 2018. PMID: 30619849 Free PMC article. Review. - Circadian control of heparan sulfate levels times phagocytosis of amyloid beta aggregates.
Clark GT, Yu Y, Urban CA, Fu G, Wang C, Zhang F, Linhardt RJ, Hurley JM. Clark GT, et al. PLoS Genet. 2022 Feb 10;18(2):e1009994. doi: 10.1371/journal.pgen.1009994. eCollection 2022 Feb. PLoS Genet. 2022. PMID: 35143487 Free PMC article.
References
- Ai X, Do AT, Kusche-Gullberg M, Lindahl U, Lu K, Emerson CP., Jr 2006. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J Biol Chem. 281:4969–4976 - PubMed
- Berninsone PM, Hirschberg CB. 2000. Nucleotide sugar transporters of the Golgi apparatus. Curr Opin Struct Biol. 10:542–547 - PubMed
- Bui C, Ouzzine M, Talhaoui I, Sharp S, Prydz K, Coughtrie MW, Fournel-Gigleux S. 2010. Epigenetics: methylation-associated repression of heparan sulfate 3-O-sulfotransferase gene expression contributes to the invasive phenotype of H-EMC-SS chondrosarcoma cells. FASEB J. 24:436–450 - PubMed
- Busse M, Feta A, Presto J, Wilen M, Gronning M, Kjellen L, Kusche-Gullberg M. 2007. Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. J Biol Chem. 282:32802–32810 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials