Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones - PubMed (original) (raw)
Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones
Gabriel Cabot et al. Antimicrob Agents Chemother. 2012 Dec.
Abstract
Recent reports have revealed the existence of widespread extensively drug-resistant (XDR) P. aeruginosa high-risk clones in health care settings, but there is still scarce information on their specific chromosomal (mutational) and acquired resistance mechanisms. Up to 20 (10.5%) of 190 bloodstream isolates collected from 10 Spanish hospitals met the XDR criteria. A representative number (15 per group) of isolates classified as multidrug-resistant (MDR) (22.6%), resistant to 1 to 2 classes (moderately resistant [modR]) (23.7%), or susceptible to all antibiotics (multiS) (43.2%) were investigated in parallel. Multilocus sequence typing (MLST) analysis revealed that all XDR isolates belonged to sequence type 175 (ST175) (n = 19) or ST111 (n = 1), both recognized as international high-risk clones. Clonal diversity was higher among the 15 MDR isolates (4 ST175, 2 ST111, and 8 additional STs) and especially high among the 15 modR (13 different STs) and multiS (14 STs) isolates. The XDR/MDR pattern in ST111 isolates correlated with the production of VIM-2, but none of the ST175 isolates produced acquired β-lactamases. In contrast, the analysis of resistance markers in 12 representative isolates (from 7 hospitals) of ST175 revealed that the XDR pattern was driven by the combination of AmpC hyperproduction, OprD inactivation (Q142X), 3 mutations conferring high-level fluoroquinolone resistance (GyrA T83I and D87N and ParC S87W), a G195E mutation in MexZ (involved in MexXY-OprM overexpression), and the production of a class 1 integron harboring the aadB gene (gentamicin and tobramycin resistance). Of particular interest, in nearly all the ST175 isolates, AmpC hyperproduction was driven by a novel AmpR-activating mutation (G154R), as demonstrated by complementation studies using an ampR mutant of PAO1. This work is the first to describe the specific resistance markers of widespread P. aeruginosa XDR high-risk clones producing invasive infections.
Figures
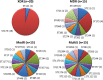
Fig 1
Distribution of STs among the XDR, MDR, modR, and multiS isolates studied. The number of isolates of each ST is shown in parentheses.
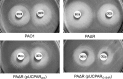
Fig 2
Results for double-disk (imipenem-ceftazidime) AmpC induction test in wild-type PAO1, the ampR knockout mutant PAΔR, and PAΔR complemented with wild-type ampR (pUCPARWT) or with the G154R mutant (pUCPARG154R).
Similar articles
- Genomics and Susceptibility Profiles of Extensively Drug-Resistant Pseudomonas aeruginosa Isolates from Spain.
Del Barrio-Tofiño E, López-Causapé C, Cabot G, Rivera A, Benito N, Segura C, Montero MM, Sorlí L, Tubau F, Gómez-Zorrilla S, Tormo N, Durá-Navarro R, Viedma E, Resino-Foz E, Fernández-Martínez M, González-Rico C, Alejo-Cancho I, Martínez JA, Labayru-Echverria C, Dueñas C, Ayestarán I, Zamorano L, Martinez-Martinez L, Horcajada JP, Oliver A. Del Barrio-Tofiño E, et al. Antimicrob Agents Chemother. 2017 Oct 24;61(11):e01589-17. doi: 10.1128/AAC.01589-17. Print 2017 Nov. Antimicrob Agents Chemother. 2017. PMID: 28874376 Free PMC article. - Spanish nationwide survey on Pseudomonas aeruginosa antimicrobial resistance mechanisms and epidemiology.
Del Barrio-Tofiño E, Zamorano L, Cortes-Lara S, López-Causapé C, Sánchez-Diener I, Cabot G, Bou G, Martínez-Martínez L, Oliver A; GEMARA-SEIMC/REIPI Pseudomonas study Group. Del Barrio-Tofiño E, et al. J Antimicrob Chemother. 2019 Jul 1;74(7):1825-1835. doi: 10.1093/jac/dkz147. J Antimicrob Chemother. 2019. PMID: 30989186 - Deciphering the Resistome of the Widespread Pseudomonas aeruginosa Sequence Type 175 International High-Risk Clone through Whole-Genome Sequencing.
Cabot G, López-Causapé C, Ocampo-Sosa AA, Sommer LM, Domínguez MÁ, Zamorano L, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Plesiat P, Oliver A. Cabot G, et al. Antimicrob Agents Chemother. 2016 Nov 21;60(12):7415-7423. doi: 10.1128/AAC.01720-16. Print 2016 Dec. Antimicrob Agents Chemother. 2016. PMID: 27736752 Free PMC article. - The increasing threat of Pseudomonas aeruginosa high-risk clones.
Oliver A, Mulet X, López-Causapé C, Juan C. Oliver A, et al. Drug Resist Updat. 2015 Jul-Aug;21-22:41-59. doi: 10.1016/j.drup.2015.08.002. Epub 2015 Aug 10. Drug Resist Updat. 2015. PMID: 26304792 Review. - Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update.
Del Barrio-Tofiño E, López-Causapé C, Oliver A. Del Barrio-Tofiño E, et al. Int J Antimicrob Agents. 2020 Dec;56(6):106196. doi: 10.1016/j.ijantimicag.2020.106196. Epub 2020 Oct 9. Int J Antimicrob Agents. 2020. PMID: 33045347 Review.
Cited by
- Draft Genome Sequence of VIM-2-Producing Multidrug-Resistant Pseudomonas aeruginosa ST175, an Epidemic High-Risk Clone.
Viedma E, Juan C, Otero JR, Oliver A, Chaves F. Viedma E, et al. Genome Announc. 2013 Apr 11;1(2):e0011213. doi: 10.1128/genomeA.00112-13. Genome Announc. 2013. PMID: 23580706 Free PMC article. - Systemic infection facilitates transmission of Pseudomonas aeruginosa in mice.
Bachta KER, Allen JP, Cheung BH, Chiu CH, Hauser AR. Bachta KER, et al. Nat Commun. 2020 Jan 28;11(1):543. doi: 10.1038/s41467-020-14363-4. Nat Commun. 2020. PMID: 31992714 Free PMC article. - Pseudomonas aeruginosa AmpR: an acute-chronic switch regulator.
Balasubramanian D, Kumari H, Mathee K. Balasubramanian D, et al. Pathog Dis. 2015 Mar;73(2):1-14. doi: 10.1111/2049-632X.12208. Epub 2015 Feb 26. Pathog Dis. 2015. PMID: 25066236 Free PMC article. Review. - Mechanisms of Resistance to Ceftolozane/Tazobactam in Pseudomonas aeruginosa: Results of the GERPA Multicenter Study.
Fournier D, Carrière R, Bour M, Grisot E, Triponney P, Muller C, Lemoine J, Jeannot K, Plésiat P; GERPA Study Group. Fournier D, et al. Antimicrob Agents Chemother. 2021 Jan 20;65(2):e01117-20. doi: 10.1128/AAC.01117-20. Print 2021 Jan 20. Antimicrob Agents Chemother. 2021. PMID: 33199392 Free PMC article. - Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections.
Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, Benito N, Grau S. Horcajada JP, et al. Clin Microbiol Rev. 2019 Aug 28;32(4):e00031-19. doi: 10.1128/CMR.00031-19. Print 2019 Sep 18. Clin Microbiol Rev. 2019. PMID: 31462403 Free PMC article. Review.
References
- Balcewich MD, et al. 2010. Crystal structure of the AmpR effector binding domain provides insight into the molecular regulation of inducible AmpC beta-lactamase. J. Mol. Biol. 400:998–1010 - PubMed
- Baquero F. 2004. From pieces to patterns: evolutionary engineering in bacterial pathogens. Nat. Rev. Microbiol. 2:510–518 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous