Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus - PubMed (original) (raw)
Comparative Study
. 2012 Oct 16;109 Suppl 2(Suppl 2):17266-72.
doi: 10.1073/pnas.1121260109. Epub 2012 Oct 8.
Affiliations
- PMID: 23045659
- PMCID: PMC3477392
- DOI: 10.1073/pnas.1121260109
Comparative Study
Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus
Matthew Suderman et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Early life experience is associated with long-term effects on behavior and epigenetic programming of the NR3C1 (GLUCOCORTICOID RECEPTOR) gene in the hippocampus of both rats and humans. However, it is unlikely that such effects completely capture the evolutionarily conserved epigenetic mechanisms of early adaptation to environment. Here we present DNA methylation profiles spanning 6.5 million base pairs centered at the NR3C1 gene in the hippocampus of humans who experienced abuse as children and nonabused controls. We compare these profiles to corresponding DNA methylation profiles in rats that received differential levels of maternal care. The profiles of both species reveal hundreds of DNA methylation differences associated with early life experience distributed across the entire region in nonrandom patterns. For instance, methylation differences tend to cluster by genomic location, forming clusters covering as many as 1 million bases. Even more surprisingly, these differences seem to specifically target regulatory regions such as gene promoters, particularly those of the protocadherin α, β, and γ gene families. Beyond these high-level similarities, more detailed analyses reveal methylation differences likely stemming from the significant biological and environmental differences between species. These results provide support for an analogous cross-species epigenetic regulatory response at the level of the genomic region to early life experience.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Associations of human DNA methylation with early life abuse in the 6.5-Mb NR3C1 locus. Track images obtained from the University of California, Santa Cruz genome browser (human genome assembly hg18) show % 5meC: average methylation levels across all samples estimated from microarray probe intensities; Δ 5meC: mean log2 fold differences between abused and control sample probe intensities, where positive values are shown in black and indicate higher methylation in abused samples, and gray values indicate higher methylation in control samples; cDMR: locations of cDMRs (significantly higher methylation in control samples); and aDMR: locations of aDMRs (significantly higher methylation in abuse samples). The locations of the protocadherin families of genes and NR3C1 are identified by shading.
Fig. 2.
Associations of human and rat DNA methylation with early life abuse in the 6.5-Mb NR3C1 locus. Top, Middle, and Bottom: Each panel shows the 6.5-Mb NR3C1 locus. Top: Human locus. Bottom: Rat locus. Middle: Human–rat “hybrid” panel created by assigning orthologous genes to positions similar to their relative locations in the human and rat genomes. Each panel is divided into five labeled parts: genes: black horizontal arrows denote genes and the direction of mRNA synthesis; variation: graph indicates regions of high and low methylation variation across all human subjects; % homology: graph shows the percentage of bases in the human genome that were mapped by the lastz alignment tool to the rat genome; 5meC diff: graph shows mean log2 fold differences between sample groups (i.e., between abused and control humans and between high- and low-LG rats); and % 5meC: graph shows methylation levels estimated from microarray probe intensities. Across each panel, gray vertical lines demark transcription start sites. Black lines between panels link the positions of transcription start sites of orthologous genes. The lack of crossings between these lines illustrates conservation of gene architecture around NR3C1 between rats and humans.
Fig. 3.
Validation of microarray calls. Real-time PCR validation of microarray meDIP data is shown. Eleven of the 28 promoters identified as being differentially methylated by microarray (
Table S1
) were subjected to real-time PCR quantification of enrichment. The y axis represents concentration values generated by methylation-enriched and input DNA. Left: Concentration levels in the abuse group. Right: Concentration levels in the control group. Each real-time PCR was performed in triplicate. Error bars indicate SEM.
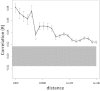
Fig. 4.
Correlation of human DNA methylation associations with early life abuse in the 6.5-Mb NR3C1 locus. Pearson correlations of DNA methylation differences between the subject groups at various genomic distances. Error bars show 95% confidence intervals for the correlation values. The gray highlight shows the expected 95% confidence interval if there is no correlation between methylation differences at different genomic sites. This confidence interval does not overlap with the error bars associated with distances less than 1 Mb, suggesting the existence of systematic dependencies between methylation differences at distances up to 1 Mb.
Similar articles
- Broad epigenetic signature of maternal care in the brain of adult rats.
McGowan PO, Suderman M, Sasaki A, Huang TC, Hallett M, Meaney MJ, Szyf M. McGowan PO, et al. PLoS One. 2011 Feb 28;6(2):e14739. doi: 10.1371/journal.pone.0014739. PLoS One. 2011. PMID: 21386994 Free PMC article. - Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse.
McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. McGowan PO, et al. Nat Neurosci. 2009 Mar;12(3):342-8. doi: 10.1038/nn.2270. Nat Neurosci. 2009. PMID: 19234457 Free PMC article. - Glucocorticoid receptor gene (NR3C1) methylation following stressful events between birth and adolescence. The TRAILS study.
van der Knaap LJ, Riese H, Hudziak JJ, Verbiest MM, Verhulst FC, Oldehinkel AJ, van Oort FV. van der Knaap LJ, et al. Transl Psychiatry. 2014 Apr 8;4(4):e381. doi: 10.1038/tp.2014.22. Transl Psychiatry. 2014. PMID: 24713862 Free PMC article. - Effects of the Social Environment and Stress on Glucocorticoid Receptor Gene Methylation: A Systematic Review.
Turecki G, Meaney MJ. Turecki G, et al. Biol Psychiatry. 2016 Jan 15;79(2):87-96. doi: 10.1016/j.biopsych.2014.11.022. Epub 2014 Dec 13. Biol Psychiatry. 2016. PMID: 25687413 Free PMC article. Review. - Early life trauma, depression and the glucocorticoid receptor gene--an epigenetic perspective.
Smart C, Strathdee G, Watson S, Murgatroyd C, McAllister-Williams RH. Smart C, et al. Psychol Med. 2015 Dec;45(16):3393-410. doi: 10.1017/S0033291715001555. Epub 2015 Sep 21. Psychol Med. 2015. PMID: 26387521 Review.
Cited by
- Functional diversity of microglia - how heterogeneous are they to begin with?
Hanisch UK. Hanisch UK. Front Cell Neurosci. 2013 May 14;7:65. doi: 10.3389/fncel.2013.00065. eCollection 2013. Front Cell Neurosci. 2013. PMID: 23717262 Free PMC article. - Omics Application in Animal Science-A Special Emphasis on Stress Response and Damaging Behaviour in Pigs.
Kasper C, Ribeiro D, Almeida AM, Larzul C, Liaubet L, Murani E. Kasper C, et al. Genes (Basel). 2020 Aug 11;11(8):920. doi: 10.3390/genes11080920. Genes (Basel). 2020. PMID: 32796712 Free PMC article. Review. - Methylation at the CpG island shore region upregulates Nr3c1 promoter activity after early-life stress.
Bockmühl Y, Patchev AV, Madejska A, Hoffmann A, Sousa JC, Sousa N, Holsboer F, Almeida OF, Spengler D. Bockmühl Y, et al. Epigenetics. 2015;10(3):247-57. doi: 10.1080/15592294.2015.1017199. Epigenetics. 2015. PMID: 25793778 Free PMC article. - Epigenetic pathways through which experiences become linked with biology.
McGowan PO, Roth TL. McGowan PO, et al. Dev Psychopathol. 2015 May;27(2):637-48. doi: 10.1017/S0954579415000206. Dev Psychopathol. 2015. PMID: 25997776 Free PMC article. Review. - DNA methylation associated with healthy aging of elderly twins.
Kim S, Wyckoff J, Morris AT, Succop A, Avery A, Duncan GE, Jazwinski SM. Kim S, et al. Geroscience. 2018 Dec;40(5-6):469-484. doi: 10.1007/s11357-018-0040-0. Epub 2018 Aug 22. Geroscience. 2018. PMID: 30136078 Free PMC article.
References
- Francis D, Diorio J, Liu D, Meaney MJ. Nongenomic transmission across generations of maternal behavior and stress responses in the rat. Science. 1999;286:1155–1158. - PubMed
- Gilbert R, et al. Burden and consequences of child maltreatment in high-income countries. Lancet. 2009;373:68–81. - PubMed
- Weaver IC, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases