Trisomy 21-associated defects in human primitive hematopoiesis revealed through induced pluripotent stem cells - PubMed (original) (raw)
. 2012 Oct 23;109(43):17573-8.
doi: 10.1073/pnas.1211175109. Epub 2012 Oct 8.
Marta Byrska-Bishop, Joanna M Tober, Yu Yao, Daniel Vandorn, Joanna B Opalinska, Jason A Mills, John Kim Choi, Nancy A Speck, Paul Gadue, Ross C Hardison, Richard L Nemiroff, Deborah L French, Mitchell J Weiss
Affiliations
- PMID: 23045704
- PMCID: PMC3491490
- DOI: 10.1073/pnas.1211175109
Trisomy 21-associated defects in human primitive hematopoiesis revealed through induced pluripotent stem cells
Stella T Chou et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Patients with Down syndrome (trisomy 21, T21) have hematologic abnormalities throughout life. Newborns frequently exhibit abnormal blood counts and a clonal preleukemia. Human T21 fetal livers contain expanded erythro-megakaryocytic precursors with enhanced proliferative capacity. The impact of T21 on the earliest stages of embryonic hematopoiesis is unknown and nearly impossible to examine in human subjects. We modeled T21 yolk sac hematopoiesis using human induced pluripotent stem cells (iPSCs). Blood progenitor populations generated from T21 iPSCs were present at normal frequency and proliferated normally. However, their developmental potential was altered with enhanced erythropoiesis and reduced myelopoiesis, but normal megakaryocyte production. These abnormalities overlap with those of T21 fetal livers, but also reflect important differences. Our studies show that T21 confers distinct developmental stage- and species-specific hematopoietic defects. More generally, we illustrate how iPSCs can provide insight into early stages of normal and pathological human development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
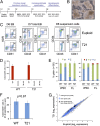
T21 iPSCs produce normal levels of early hematopoietic progenitors. (A) Schematic of hematopoietic differentiation protocol via EB formation with the following cytokines: BMP4, VEGF, SCF, TPO, FLT3, bFGF, EPO, IL-3, IL-11, and IGF1. (B) Photograph of iPSC-derived EB culture with hematopoietic cells released into the medium. Original magnification, 10×. (C) Flow cytometry analysis showing CD31+KDR+ hematoendothelial precursor cells (Left) and CD34+/−43+235+41+ progenitors within EBs (Center), and released into the medium (Right). (D) Methylcellulose colony assays of various purified populations. Cytokines include SCF, IL-3, EPO, and GMCSF. Results show mean values ± SEM, n = 3 per group. (E) Globin gene expression in erythroid colonies from iPSCs or fetal liver (FL) determined by quantitative real-time PCR. Charts show fraction of α- (Left) or β-like (Right) genes. Ten to 20 colonies were pooled per sample, n = 3 per group. (F) Frequency of CD43+41+235+ progenitor cells in EB cultures on day 7–8 of hematopoietic differentiation (n = 14 and 11 independent experiments for euploid and T21 iPSCs, respectively. (G) Scatter plots of microarray data showing average mRNA expression values in purified CD43+41+235+ progenitors from between-group comparison of three euploid and three T21 biological replicate samples.
Fig. 2.
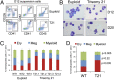
Propensity for erythroid differentiation by T21 iPSCs. (A) Flow cytometry analysis of suspension cells in day 12 differentiation cultures showing mature hematopoietic lineages: erythroid (Ery, CD41−235+), megakaryocytic (Meg, CD41+42+), and myeloid (CD45+18+). (B) May–Grunwald Giemsa-stained cells from EB suspension cultures at days 12 and 20. (Scale bars, 50 μm.) (C) Distribution of lineage-committed cells in EB suspension cultures at days 12–14 of differentiation. n = 3–5 independent experiments per iPSC line. (D) Summary of data with all iPSC lines combined according to genotype (n = 15 per group).
Fig. 3.
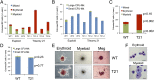
Increased erythroid progenitors in T21 iPSC differentiation cultures. CD43+41+235+ hematopoietic progenitors derived from three euploid and four T21 iPSC lines were analyzed. (A) Methylcellulose colony assays of CD43+41+235+ progenitors containing SCF, IL-3, EPO, GM-CSF, and (B) Colony-forming megakaryocyte (CFU-Mk) assays that include TPO, IL-3, and IL-6. Results show mean values ± SEM for three independent experiments per iPSC line. (C) Summary of data with all iPSC lines combined according to genotype for methylcellulose colony assays and (D) CFU-Mk assays. Results show mean values ± SEM, n = 9 for WT, n = 12 for T21. (E) Representative hematopoietic colonies from iPSC-derived progenitors. (Scale bars, 200 μm.) Meg, megakaryocyte. (F) Morphology of cells from iPSC-derived erythroid and myeloid colonies. May–Grunwald Giemsa stain. (Scale bars, 20 μm.)
Comment in
- Leukaemia: Early changes.
McCarthy N. McCarthy N. Nat Rev Cancer. 2012 Dec;12(12):799. doi: 10.1038/nrc3403. Epub 2012 Nov 15. Nat Rev Cancer. 2012. PMID: 23151601 No abstract available.
Similar articles
- Altered hematopoiesis in trisomy 21 as revealed through in vitro differentiation of isogenic human pluripotent cells.
Maclean GA, Menne TF, Guo G, Sanchez DJ, Park IH, Daley GQ, Orkin SH. Maclean GA, et al. Proc Natl Acad Sci U S A. 2012 Oct 23;109(43):17567-72. doi: 10.1073/pnas.1215468109. Epub 2012 Oct 8. Proc Natl Acad Sci U S A. 2012. PMID: 23045682 Free PMC article. - Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21.
Roy A, Cowan G, Mead AJ, Filippi S, Bohn G, Chaidos A, Tunstall O, Chan JK, Choolani M, Bennett P, Kumar S, Atkinson D, Wyatt-Ashmead J, Hu M, Stumpf MP, Goudevenou K, O'Connor D, Chou ST, Weiss MJ, Karadimitris A, Jacobsen SE, Vyas P, Roberts I. Roy A, et al. Proc Natl Acad Sci U S A. 2012 Oct 23;109(43):17579-84. doi: 10.1073/pnas.1211405109. Epub 2012 Oct 8. Proc Natl Acad Sci U S A. 2012. PMID: 23045701 Free PMC article. - Single-cell transcriptomics reveal individual and synergistic effects of Trisomy 21 and GATA1s on hematopoiesis.
Takasaki K, Wafula EK, Kumar SS, Smith D, Sit YT, Gagne AL, French DL, Thom CS, Chou ST. Takasaki K, et al. bioRxiv [Preprint]. 2024 Oct 31:2024.05.24.595827. doi: 10.1101/2024.05.24.595827. bioRxiv. 2024. PMID: 38826323 Free PMC article. Preprint. - The impact of trisomy 21 on foetal haematopoiesis.
Roberts I, O'Connor D, Roy A, Cowan G, Vyas P. Roberts I, et al. Blood Cells Mol Dis. 2013 Dec;51(4):277-81. doi: 10.1016/j.bcmd.2013.07.008. Epub 2013 Aug 7. Blood Cells Mol Dis. 2013. PMID: 23932236 Free PMC article. Review. - Cellular Basis of Embryonic Hematopoiesis and Its Implications in Prenatal Erythropoiesis.
Yamane T. Yamane T. Int J Mol Sci. 2020 Dec 8;21(24):9346. doi: 10.3390/ijms21249346. Int J Mol Sci. 2020. PMID: 33302450 Free PMC article. Review.
Cited by
- Down syndrome-associated leukaemias: current evidence and challenges.
Mason NR, Cahill H, Diamond Y, McCleary K, Kotecha RS, Marshall GM, Mateos MK. Mason NR, et al. Ther Adv Hematol. 2024 Jul 23;15:20406207241257901. doi: 10.1177/20406207241257901. eCollection 2024. Ther Adv Hematol. 2024. PMID: 39050114 Free PMC article. Review. - Immune cells and RBCs derived from human induced pluripotent stem cells: method, progress, prospective challenges.
Jiang JH, Ren RT, Cheng YJ, Li XX, Zhang GR. Jiang JH, et al. Front Cell Dev Biol. 2024 Jan 5;11:1327466. doi: 10.3389/fcell.2023.1327466. eCollection 2023. Front Cell Dev Biol. 2024. PMID: 38250324 Free PMC article. Review. - Synergistic roles of DYRK1A and GATA1 in trisomy 21 megakaryopoiesis.
Sit YT, Takasaki K, An HH, Xiao Y, Hurtz C, Gearhart PA, Zhang Z, Gadue P, French DL, Chou ST. Sit YT, et al. JCI Insight. 2023 Oct 31;8(23):e172851. doi: 10.1172/jci.insight.172851. JCI Insight. 2023. PMID: 37906251 Free PMC article. - Generation of Red Blood Cells from Human Pluripotent Stem Cells-An Update.
Lee SJ, Jung C, Oh JE, Kim S, Lee S, Lee JY, Yoon YS. Lee SJ, et al. Cells. 2023 Jun 5;12(11):1554. doi: 10.3390/cells12111554. Cells. 2023. PMID: 37296674 Free PMC article. Review. - Bullous eruptions in transient abnormal myelopoiesis with normal phenotype.
Shivamallappa MD, Mullins A, Browning Carmo K. Shivamallappa MD, et al. BMJ Case Rep. 2023 Apr 7;16(4):e251523. doi: 10.1136/bcr-2022-251523. BMJ Case Rep. 2023. PMID: 37028822
References
- Hassold T, Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–291. - PubMed
- Henry E, Walker D, Wiedmeier SE, Christensen RD. Hematological abnormalities during the first week of life among neonates with Down syndrome: Data from a multihospital healthcare system. Am J Med Genet A. 2007;143(1):42–50. - PubMed
- Roizen NJ, Amarose AP. Hematologic abnormalities in children with Down syndrome. Am J Med Genet. 1993;46(5):510–512. - PubMed
- Roy A, Roberts I, Norton A, Vyas P. Acute megakaryoblastic leukaemia (AMKL) and transient myeloproliferative disorder (TMD) in Down syndrome: A multi-step model of myeloid leukaemogenesis. Br J Haematol. 2009;147(1):3–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK090969/DK/NIDDK NIH HHS/United States
- RC2 HG005573/HG/NHGRI NIH HHS/United States
- R01 HL091724/HL/NHLBI NIH HHS/United States
- F32 HL010166/HL/NHLBI NIH HHS/United States
- R01 DK065806/DK/NIDDK NIH HHS/United States
- K08 HL093290/HL/NHLBI NIH HHS/United States
- RC2 HL10166/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical