Combinatorial regulation of tissue specification by GATA and FOG factors - PubMed (original) (raw)
Review
Combinatorial regulation of tissue specification by GATA and FOG factors
Timothy M Chlon et al. Development. 2012 Nov.
Abstract
The development of complex organisms requires the formation of diverse cell types from common stem and progenitor cells. GATA family transcriptional regulators and their dedicated co-factors, termed Friend of GATA (FOG) proteins, control cell fate and differentiation in multiple tissue types from Drosophila to man. FOGs can both facilitate and antagonize GATA factor transcriptional regulation depending on the factor, cell, and even the specific gene target. In this review, we highlight recent studies that have elucidated mechanisms by which FOGs regulate GATA factor function and discuss how these factors use these diverse modes of gene regulation to control cell lineage specification throughout metazoans.
Figures
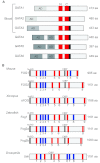
Fig. 1.
Conservation of FOG proteins throughout evolution. (A) The six mammalian GATA factors, which are well conserved throughout vertebrates. Each GATA factor contains two highly conserved zinc finger domains (N and C, red), a conserved nuclear localization signal (NLS) that lies within a basic region C-terminal to the C-finger (black), and a poorly conserved N-terminal activation domain (AD). The shading of the N-terminal activation domains indicates the divergence in sequence of this domain. (B) FOG family members in mouse, frog, zebrafish and fruit fly. Zinc finger domains are represented by colored vertical bars: red, C2HC; blue, C2H2. The black bars indicate the presence of a conserved co-repressor interaction motif (NuRD or CtBP). The sizes and distributions of the zinc fingers are approximations. The question mark in zebrafish Fog1 indicates the presence of a predicted zinc finger domain that is not conserved in mammalian FOG proteins.
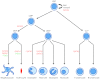
Fig. 2.
Control of mammalian hematopoiesis by GATA and FOG factors. A classical representation of mammalian hematopoiesis is depicted, and the roles of GATA and FOG proteins at each step are indicated. HSC, hematopoietic stem cell; CMP, common myeloid progenitor; MEP, megakaryocyte-erythrocyte progenitor; GMP, granulocyte/macrophage progenitor; CLP, common lymphoid progenitor.
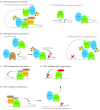
Fig. 3.
FOG1 controls GATA1 transcriptional activity through diverse mechanisms. (A) FOG1 is required for activation of a subset of GATA1 target genes, including (i) β-globin and (ii) Band3. At the β-globin gene, GATA1 and FOG1 occupy the locus control region (LCR), recruit the SCL and NuRD complexes, and participate in a chromatin loop that brings the LCR into close proximity with the β-globin promoter. At the Band3 gene, FOG1 facilitates GATA1 chromatin occupancy to induce gene activation. (B) FOG1 is required for repression of a subset of GATA1 target genes, including (i) Gata2 and (ii) Kit. At Gata2, GATA1 and FOG1 eject GATA2 from cis-regulatory elements (GATA switch), and FOG1 recruits the NuRD repressor to induce locus-wide histone deacetylation. At Kit, FOG1 is required for GATA1 to alter chromatin loops involving distal enhancer elements. (C) A subset of GATA1-activated genes, including Fog1 itself, do not require FOG1. (D) A subset of GATA1-repressed genes, including Lyl1, do not require FOG1. (E) FOG1 inhibits GATA1 activation of some target genes. In some cases, FOG1 accomplishes this regulation by prohibiting GATA1 chromatin occupancy. Several mast cell-specific genes, including Fcer1b (Ms4a2), are regulated by this mechanism.
Similar articles
- GATA proteins work together with friend of GATA (FOG) and C-terminal binding protein (CTBP) co-regulators to control adipogenesis.
Jack BH, Crossley M. Jack BH, et al. J Biol Chem. 2010 Oct 15;285(42):32405-14. doi: 10.1074/jbc.M110.141317. Epub 2010 Aug 11. J Biol Chem. 2010. PMID: 20705609 Free PMC article. - The Friend of GATA proteins U-shaped, FOG-1, and FOG-2 function as negative regulators of blood, heart, and eye development in Drosophila.
Fossett N, Tevosian SG, Gajewski K, Zhang Q, Orkin SH, Schulz RA. Fossett N, et al. Proc Natl Acad Sci U S A. 2001 Jun 19;98(13):7342-7. doi: 10.1073/pnas.131215798. Epub 2001 Jun 12. Proc Natl Acad Sci U S A. 2001. PMID: 11404479 Free PMC article. - Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage.
Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR, Akashi K, Orkin SH. Cantor AB, et al. J Exp Med. 2008 Mar 17;205(3):611-24. doi: 10.1084/jem.20070544. Epub 2008 Feb 25. J Exp Med. 2008. PMID: 18299398 Free PMC article. - Navigating Transcriptional Coregulator Ensembles to Establish Genetic Networks: A GATA Factor Perspective.
DeVilbiss AW, Tanimura N, McIver SC, Katsumura KR, Johnson KD, Bresnick EH. DeVilbiss AW, et al. Curr Top Dev Biol. 2016;118:205-44. doi: 10.1016/bs.ctdb.2016.01.003. Epub 2016 Feb 23. Curr Top Dev Biol. 2016. PMID: 27137658 Review. - Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins.
Cantor AB, Orkin SH. Cantor AB, et al. Semin Cell Dev Biol. 2005 Feb;16(1):117-28. doi: 10.1016/j.semcdb.2004.10.006. Epub 2004 Dec 15. Semin Cell Dev Biol. 2005. PMID: 15659346 Review.
Cited by
- Transcriptional control of chondrocyte specification and differentiation.
Liu CF, Samsa WE, Zhou G, Lefebvre V. Liu CF, et al. Semin Cell Dev Biol. 2017 Feb;62:34-49. doi: 10.1016/j.semcdb.2016.10.004. Epub 2016 Oct 19. Semin Cell Dev Biol. 2017. PMID: 27771362 Free PMC article. Review. - The role of genes and environment in the etiology of congenital diaphragmatic hernias.
Burns NG, Kardon G. Burns NG, et al. Curr Top Dev Biol. 2023;152:115-138. doi: 10.1016/bs.ctdb.2022.10.004. Epub 2022 Nov 23. Curr Top Dev Biol. 2023. PMID: 36707209 Free PMC article. Review. - Combinatorial rules of precursor specification underlying olfactory neuron diversity.
Li Q, Ha TS, Okuwa S, Wang Y, Wang Q, Millard SS, Smith DP, Volkan PC. Li Q, et al. Curr Biol. 2013 Dec 16;23(24):2481-90. doi: 10.1016/j.cub.2013.10.053. Epub 2013 Nov 21. Curr Biol. 2013. PMID: 24268416 Free PMC article. - The Molecular Signature of Megakaryocyte-Erythroid Progenitors Reveals a Role for the Cell Cycle in Fate Specification.
Lu YC, Sanada C, Xavier-Ferrucio J, Wang L, Zhang PX, Grimes HL, Venkatasubramanian M, Chetal K, Aronow B, Salomonis N, Krause DS. Lu YC, et al. Cell Rep. 2018 Nov 20;25(8):2083-2093.e4. doi: 10.1016/j.celrep.2018.10.084. Cell Rep. 2018. PMID: 30463007 Free PMC article. - Haemopedia: An Expression Atlas of Murine Hematopoietic Cells.
de Graaf CA, Choi J, Baldwin TM, Bolden JE, Fairfax KA, Robinson AJ, Biben C, Morgan C, Ramsay K, Ng AP, Kauppi M, Kruse EA, Sargeant TJ, Seidenman N, D'Amico A, D'Ombrain MC, Lucas EC, Koernig S, Baz Morelli A, Wilson MJ, Dower SK, Williams B, Heazlewood SY, Hu Y, Nilsson SK, Wu L, Smyth GK, Alexander WS, Hilton DJ. de Graaf CA, et al. Stem Cell Reports. 2016 Sep 13;7(3):571-582. doi: 10.1016/j.stemcr.2016.07.007. Epub 2016 Aug 4. Stem Cell Reports. 2016. PMID: 27499199 Free PMC article.
References
- Abel T., Michelson A. M., Maniatis T. (1993). A Drosophila GATA family member that binds to Adh regulatory sequences is expressed in the developing fat body. Development 119, 623-633 - PubMed
- Anttonen M., Ketola I., Parviainen H., Pusa A.-K., Heikinheimo M. (2003). FOG-2 and GATA-4 are coexpressed in the mouse ovary and can modulate mullerian-inhibiting substance expression. Biol. Reprod. 68, 1333-1340 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases