The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization - PubMed (original) (raw)
The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization
Rui Jia et al. J Virol. 2012 Dec.
Abstract
Interferon-inducible transmembrane (IFITM) protein family members IFITM1, -2, and -3 restrict the infection of multiple enveloped viruses. Significant enrichment of a minor IFITM3 allele was recently reported for patients who were hospitalized for seasonal and 2009 H1N1 pandemic flu. This IFITM3 allele lacks the region corresponding to the first amino-terminal 21 amino acids and is unable to inhibit influenza A virus. In this study, we found that deleting this 21-amino-acid region relocates IFITM3 from the endosomal compartments to the cell periphery. This finding likely underlies the lost inhibition of influenza A virus that completes its entry exclusively within endosomes at low pH. Yet, wild-type IFITM3 and the mutant with the 21-amino-acid deletion inhibit HIV-1 replication equally well. Given the pH-independent nature of HIV-1 entry, our results suggest that IFITM3 can inhibit viruses that enter cells via different routes and that its N-terminal region is specifically required for controlling pH-dependent viruses.
Figures
Fig 1
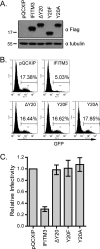
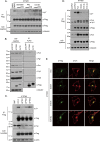
IFITM3 inhibits infection by VSV-G protein-pseudotyped viruses. (A) Illustration of IFITM3 and its N-terminal mutations. The two transmembrane domains (TM1 and TM2) and the N-terminal sequence of IFITM3 are depicted. Sequences that are mutated in each mutant are also shown. (B) Western blotting to show the expression of IFITM3 and its mutants in stably transduced HEK293 cells. Tubulin was probed as an internal control. (C) Flow cytometry data of one representative infection experiment showing the effect of IFITM3 and its mutants on infection by VSV-G protein-pseudotyped MLV reporter viruses. Control cells were stably transduced with the pQCXIP empty retroviral vector. The percentages of infected cells (GFP positive) are indicated. (D) The relative infectivity was calculated by dividing the percentages of GFP-positive cells from IFITM3-expressing cell lines by the percentage from the control cell line (pQCXIP). The results are the averages of three independent infection experiments. The error bars indicate standard deviations.
Fig 2
Mutating Y20 abolishes IFITM3 inhibition of infection by VSV-G protein-pseudotyped virus. (A) Expression of the Y20A and Y20F mutants in stably transduced HEK293 cells. (B) Infection of HEK293 cell lines by VSV-G protein-pseudotyped MLV reporter viruses. Results shown are from one representative infection. (C) Summary of three independent infections. The infection efficiency of the control cells is arbitrarily set as 1.
Fig 3
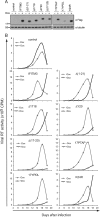
IFITM3 and its N-terminal mutants suppress HIV-1 replication. (A) IFITM3 and its various mutants were cloned into the pRetro-X-Tight vector and stably transduced into SupT1 cells. Expression of IFITM3 and its mutants was induced by doxycycline (Dox) and examined by Western blotting. (B) HIV-1 replication. SupT1 cells were infected with HIV-1. Virus growth was monitored over various time periods through measuring viral reverse transcriptase activity in the culture supernatants.
Fig 4
Effects of IFITM3 and the Δ(1–21) mutant on virus entry. (A) BlaM-Vpr-labeled BH10 viruses were used to infect SupT1 cells that were stably transduced with the pRetro-X-Tight vector itself (control) or the vector expressing IFITM3 or the Δ(1–21) mutant. The number of infected cells was determined by monitoring the cleavage of the CCF2/AM substrate by BlaM-Vpr. (B) Results of similar experiments performed with BlaM-Vpr labeled NLENY1-ES-IRES viruses that were pseudotyped with VSV-G protein. Data shown are the averages of three independent infections.
Fig 5
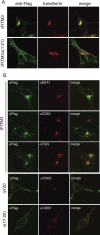
Cellular location of IFITM3 and its mutants. (A) IFITM3 and Δ(1–21) mutant DNAs were transfected into HEK293 cells. Prior to fixation, cells were incubated with Alexa Fluor 555-conjugated transferrin (pseudocolored in red) (5 μg/ml in serum-free Dulbecco modified Eagle medium) for 10 min at 37°C. IFITM3 and the Δ(1–21) mutant were detected by immunostaining with anti-Flag antibodies (pseudocolored in green). (B) IFITM3 and Δ(17–20) and ΔY20 mutant DNAs were transfected into HEK293 cells. Their expression was detected by immunostaining with anti-Flag antibodies (pseudocolored in green). The locations of EEA1, CD63, and TGN46 were visualized by immunostaining with respective antibodies (pseudocolored in red). Representative images are shown.
Fig 6
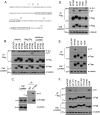
IFITM3 is phosphorylated at Y20. (A) Illustration of IFITM3 sequence and the location of tyrosine residues. (B) Detection of IFITM3 phosphorylation using antibodies recognizing phosphorylated tyrosine (pY). DNA vectors expressing IFITM1, IFITM2, and IFITM3 were transfected into HEK293T cells. Na3VO4 or phosphatase inhibitor cocktail was used to treat cells before cell lysates were harvested. The IFITM1, IFITM2, and IFITM3 proteins were precipitated (IP) with anti-Flag antibodies and then tested by Western blotting with anti-pY antibodies. (C) Phosphorylation of endogenous IFITM3. HeLa cells were treated with 500 U/ml IFN-α for 24 h, followed by incubation with 10 mM Na3VO4 for 1 h prior to harvest. The endogenous IFITM3 was precipitated and subject to Western blotting with anti-pY antibody. (D) Detection of phosphorylation of wild-type IFITM3 and the ΔY20, Y80A, Y99A, and Y132A mutants in Western blotting with anti-pY antibodies. (E) Tyrosine phosphorylation of wild-type IFITM3 and theΔY20, Y20F, and Y20A mutants. (F) Tyrosine phosphorylation of IFITM3 and the Δ(1–21), Δ1718, ΔY20, Δ(17–20), 17PTAP, 17YPDL, and K24R mutants. Positions of the IgG light chain (Ig-L) on the gels are indicated by arrows.
Fig 7
Identification of tyrosine kinase that phosphorylates IFITM3. (A) Effects of kinase inhibitors on IFITM3 phosphorylation. Cells were treated with genistein, PP2, or U0126 to inhibit Src family kinases or the ERK family. Phosphorylation of IFITM3 was detected with antibodies against pY. (B) Interaction of IFITM3 with Src family members, including Fyn, Fgr, Src, Lyn, and Hck. Wild-type IFITM3 or its ΔY20 mutant was precipitated with anti-Flag antibodies and then assessed by Western blotting with antibodies recognizing Fyn, Fgr, Src, Lyn, or Hck. (C) Effect of Fyn knockdown on IFITM3 phosphorylation. HEK293T cells were transfected with siRNA oligonucleotides targeting Fyn and IFITM3 mRNAs. Phosphorylation of IFITM3 was then evaluated by Western blotting using anti-pY antibodies. (D) Effects of Fyn and its GGAA, E314A, and E314Q mutants on IFITM3 phosphorylation. Wild-type Fyn or these mutants were transiently expressed in HEK293T cells that had also been transfected with IFITM3 plasmid DNA. The ectopically expressed IFITM3 was immunoprecipitated with anti-Flag antibody and then subjected to Western blotting for the detection of Fyn and its mutants, as well as tyrosine-phosphorylated IFITM3. The position of the IgG heavy chain (Ig-H) on the gel is indicated by an arrow. (E) Colocalization of Fyn with IFITM3. IFITM3, wild-type Fyn, and Fyn mutants were transiently expressed in HEK293 cells. Flag-tagged IFITM3 was detected by immunofluorescence staining with anti-Flag antibody (pseudocolored in green). Fyn and its mutants were detected with anti-Fyn antibody (pseudocolored in red). Representative images are shown.
Similar articles
- Duck Interferon-Inducible Transmembrane Protein 3 Mediates Restriction of Influenza Viruses.
Blyth GA, Chan WF, Webster RG, Magor KE. Blyth GA, et al. J Virol. 2015 Oct 14;90(1):103-16. doi: 10.1128/JVI.01593-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26468537 Free PMC article. - IFITM3 restricts the morbidity and mortality associated with influenza.
Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, Wise HM, Kane L, Goulding D, Digard P, Anttila V, Baillie JK, Walsh TS, Hume DA, Palotie A, Xue Y, Colonna V, Tyler-Smith C, Dunning J, Gordon SB; GenISIS Investigators; MOSAIC Investigators; Smyth RL, Openshaw PJ, Dougan G, Brass AL, Kellam P. Everitt AR, et al. Nature. 2012 Mar 25;484(7395):519-23. doi: 10.1038/nature10921. Nature. 2012. PMID: 22446628 Free PMC article. - IFITM Proteins That Restrict the Early Stages of Respiratory Virus Infection Do Not Influence Late-Stage Replication.
Meischel T, Fritzlar S, Villalon-Letelier F, Tessema MB, Brooks AG, Reading PC, Londrigan SL. Meischel T, et al. J Virol. 2021 Sep 27;95(20):e0083721. doi: 10.1128/JVI.00837-21. Epub 2021 Jul 28. J Virol. 2021. PMID: 34319159 Free PMC article. - Regulation of the trafficking and antiviral activity of IFITM3 by post-translational modifications.
Chesarino NM, McMichael TM, Yount JS. Chesarino NM, et al. Future Microbiol. 2014;9(10):1151-63. doi: 10.2217/fmb.14.65. Future Microbiol. 2014. PMID: 25405885 Free PMC article. Review. - Discovery and Characterization of IFITM _S_-Palmitoylation.
Das T, Hang HC. Das T, et al. Viruses. 2023 Nov 28;15(12):2329. doi: 10.3390/v15122329. Viruses. 2023. PMID: 38140570 Free PMC article. Review.
Cited by
- IFITM1 is a host restriction factor that inhibits porcine epidemic diarrhea virus infection.
Cheng J, He J, Feng S, Tan L, Bai B, Dong W, Li B, Wen L, Wang A, Yuan X. Cheng J, et al. J Nanobiotechnology. 2024 Nov 5;22(1):677. doi: 10.1186/s12951-024-02884-9. J Nanobiotechnology. 2024. PMID: 39501328 Free PMC article. - Research Progress into the Biological Functions of IFITM3.
Xie Q, Wang L, Liao X, Huang B, Luo C, Liao G, Yuan L, Liu X, Luo H, Shu Y. Xie Q, et al. Viruses. 2024 Sep 29;16(10):1543. doi: 10.3390/v16101543. Viruses. 2024. PMID: 39459876 Free PMC article. Review. - Host Innate Antiviral Response to Influenza A Virus Infection: From Viral Sensing to Antagonism and Escape.
An W, Lakhina S, Leong J, Rawat K, Husain M. An W, et al. Pathogens. 2024 Jul 3;13(7):561. doi: 10.3390/pathogens13070561. Pathogens. 2024. PMID: 39057788 Free PMC article. Review. - The Antiviral Activity of Interferon-Induced Transmembrane Proteins and Virus Evasion Strategies.
Wang J, Luo Y, Katiyar H, Liang C, Liu Q. Wang J, et al. Viruses. 2024 May 6;16(5):734. doi: 10.3390/v16050734. Viruses. 2024. PMID: 38793616 Free PMC article. Review. - Role of Viral Envelope Proteins in Determining Susceptibility of Viruses to IFITM Proteins.
Marceau T, Braibant M. Marceau T, et al. Viruses. 2024 Feb 5;16(2):254. doi: 10.3390/v16020254. Viruses. 2024. PMID: 38400030 Free PMC article. Review.
References
- Akiyama T, et al. 1987. Genistein, a specific inhibitor of tyrosine-specific protein kinases. J. Biol. Chem. 262:5592–5595 - PubMed
- Alland L, Peseckis SM, Atherton RE, Berthiaume L, Resh MD. 1994. Dual myristylation and palmitylation of Src family member p59fyn affects subcellular localization. J. Biol. Chem. 269:16701–16705 - PubMed
- Bonifacino JS, Traub LM. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447 - PubMed
- Cavrois M, De Noronha C, Greene WC. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:1151–1154 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases