Competing endogenous RNA database - PubMed (original) (raw)
Competing endogenous RNA database
Aaron L Sarver et al. Bioinformation. 2012.
Abstract
A given mRNA can be regulated by interactions with miRNAs and in turn the availability of these miRNAs can be regulated by their interactions with alternate mRNAs. The concept of regulation of a given mRNA by alternate mRNA (competing endogenous mRNA) by virtue of interactions with miRNAs through shared miRNA response elements is poised to become a fundamental genetic regulatory mechanism. The molecular basis of the mRNA-mRNA cross talks is via miRNA response elements, which can be predicted based on both molecular interaction and evolutionary conservation. By examining the co-occurrence of miRNA response elements in the mRNAs on a genome-wide basis we predict competing endogenous RNA for specific mRNAs targeted by miRNAs. Comparison of the mRNAs predicted to regulate PTEN with recently published work, indicate that the results presented within the competing endogenous RNA database (ceRDB) have biological relevance.
Availability: http://www.oncomir.umn.edu/cefinder/
Keywords: MRE; ceRDB; ceRNAs; competing endogenous RNAs database; database; microRNA response elements.
Figures
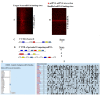
Figure 1
Visualization of co-occurrence in predicted miRNA-mRNA interactions. Heat map showing the presence of predicted miRNA-binding sites on the X-axis and the genes that contain the binding sites in the 3'UTR on the Y-axis. Only genes that show more than 5 binding sites are shown for (A) predicted interactions and (B) predicted interactions after shuffling. (C) Predicting competing mRNA via miRNA-mRNA interactions. miRNA binding site predictions in the 3'UTR are shown as colored boxes. The ‘Score’ is generated by counting the number of conserved predicted interactions. In this hypothetical case shown there are 7 predicted binding elements in the 3'UTR of the gene. (D) To predict potential competing RNA for the gene shown in A, binding sites for the predicted miRNA found in A are obtained and summed in all genes. The genes are then sorted by total number of overlapping binding sites and returned to the user. (E) Example of competing mRNA predictions from ceRDB for PTEN. The user selects an mRNA of interest from the list of available mRNA. In the case shown here the PTEN tumor suppressor is chosen. (F) Starting with the list of miRNA binding elements present in PTEN the tool predicts potential competing RNA and visualizes the extent of overlap between the miRNA binding sites. Only a representative subset of the matrix is shown, the full matrix is available online. Each predicted gene is linked back to the TargetScan database to visualize the position and total numbers of each miRNA element.
Similar articles
- lnCeDB: database of human long noncoding RNA acting as competing endogenous RNA.
Das S, Ghosal S, Sen R, Chakrabarti J. Das S, et al. PLoS One. 2014 Jun 13;9(6):e98965. doi: 10.1371/journal.pone.0098965. eCollection 2014. PLoS One. 2014. PMID: 24926662 Free PMC article. - Competing endogenous RNA: the key to posttranscriptional regulation.
Sen R, Ghosal S, Das S, Balti S, Chakrabarti J. Sen R, et al. ScientificWorldJournal. 2014 Feb 2;2014:896206. doi: 10.1155/2014/896206. eCollection 2014. ScientificWorldJournal. 2014. PMID: 24672386 Free PMC article. Review. - Interferon-alpha competing endogenous RNA network antagonizes microRNA-1270.
Kimura T, Jiang S, Yoshida N, Sakamoto R, Nishizawa M. Kimura T, et al. Cell Mol Life Sci. 2015 Jul;72(14):2749-61. doi: 10.1007/s00018-015-1875-5. Epub 2015 Mar 7. Cell Mol Life Sci. 2015. PMID: 25746225 Free PMC article. - Competing endogenous RNA networks and gastric cancer.
Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Guo LL, et al. World J Gastroenterol. 2015 Nov 7;21(41):11680-7. doi: 10.3748/wjg.v21.i41.11680. World J Gastroenterol. 2015. PMID: 26556995 Free PMC article. Review. - Competing endogenous RNA: A novel posttranscriptional regulatory dimension associated with the progression of cancer.
Dai Q, Li J, Zhou K, Liang T. Dai Q, et al. Oncol Lett. 2015 Nov;10(5):2683-2690. doi: 10.3892/ol.2015.3698. Epub 2015 Sep 14. Oncol Lett. 2015. PMID: 26722227 Free PMC article.
Cited by
- Epigenetic pathways and glioblastoma treatment.
Clarke J, Penas C, Pastori C, Komotar RJ, Bregy A, Shah AH, Wahlestedt C, Ayad NG. Clarke J, et al. Epigenetics. 2013 Aug;8(8):785-95. doi: 10.4161/epi.25440. Epub 2013 Jun 27. Epigenetics. 2013. PMID: 23807265 Free PMC article. Review. - The complexity of miRNA-mediated repression.
Wilczynska A, Bushell M. Wilczynska A, et al. Cell Death Differ. 2015 Jan;22(1):22-33. doi: 10.1038/cdd.2014.112. Epub 2014 Sep 5. Cell Death Differ. 2015. PMID: 25190144 Free PMC article. Review. - Competing Endogenous RNAs, Non-Coding RNAs and Diseases: An Intertwined Story.
Ala U. Ala U. Cells. 2020 Jun 28;9(7):1574. doi: 10.3390/cells9071574. Cells. 2020. PMID: 32605220 Free PMC article. Review. - The multilayered complexity of ceRNA crosstalk and competition.
Tay Y, Rinn J, Pandolfi PP. Tay Y, et al. Nature. 2014 Jan 16;505(7483):344-52. doi: 10.1038/nature12986. Nature. 2014. PMID: 24429633 Free PMC article. Review. - Crosstalk in competing endogenous RNA network reveals the complex molecular mechanism underlying lung cancer.
Jin X, Guan Y, Sheng H, Liu Y. Jin X, et al. Oncotarget. 2017 Aug 24;8(53):91270-91280. doi: 10.18632/oncotarget.20441. eCollection 2017 Oct 31. Oncotarget. 2017. PMID: 29207642 Free PMC article.
References
- M Van Kouwenhove, et al. Nat Rev Cancer. 2011;11:644. - PubMed
- H Guo, et al. Nature. 2010;466:835. - PubMed
- FA Karreth, et al. Cell. 2011;147:382. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials