Crystal structure of ATV(ORF273), a new fold for a thermo- and acido-stable protein from the Acidianus two-tailed virus - PubMed (original) (raw)
Crystal structure of ATV(ORF273), a new fold for a thermo- and acido-stable protein from the Acidianus two-tailed virus
Catarina Felisberto-Rodrigues et al. PLoS One. 2012.
Abstract
Acidianus two-tailed virus (ATV) infects crenarchaea of the genus Acidianus living in terrestrial thermal springs at extremely high temperatures and low pH. ATV is a member of the Bicaudaviridae virus family and undergoes extra-cellular development of two tails, a process that is unique in the viral world. To understand this intriguing phenomenon, we have undertaken structural studies of ATV virion proteins and here we present the crystal structure of one of these proteins, ATV(ORF273). ATV(ORF273) forms tetramers in solution and a molecular envelope is provided for the tetramer, computed from small-angle X-ray scattering (SAXS) data. The crystal structure has properties typical of hyperthermostable proteins, including a relatively high number of salt bridges. However, the protein also exhibits flexible loops and surface pockets. Remarkably, ATV(ORF273) displays a new α + β protein fold, consistent with the absence of homologues of this protein in public sequence databases.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
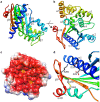
Figure 1. Crystal structure of ATV
 . a) Ribbon representation of the structure of a monomer of ATV
. a) Ribbon representation of the structure of a monomer of ATV . Its
. Its  -strands are labelled. The secondary structure elements are colored from the N′- (blue) to the C′-terminus (red). The loops that were not modelled are represented by dashed, black lines. b) A view orthogonal to the previous one, showing the arrangement of the
-strands are labelled. The secondary structure elements are colored from the N′- (blue) to the C′-terminus (red). The loops that were not modelled are represented by dashed, black lines. b) A view orthogonal to the previous one, showing the arrangement of the  -helices. This view shows part of the concave face of the monomer (top, right) c) The solvent-accessible surface of the ATV
-helices. This view shows part of the concave face of the monomer (top, right) c) The solvent-accessible surface of the ATV monomer concave face, colored by its electrostatic potential (red: −52 mV, blue: 52 mV), calculated at pH 7 and 150 mM NaCl with APBS . Two cavities are marked with yellow open stars. d) A detail view of monomer B showing the salt bridge network formed by residues Glu29/Asp33/Lys260 as well as the disulphide bond established by cysteine residues 250 and 263. Amino acid residues are labelled in one-letter code; the secondary structure elements to which they belong are also labelled.
monomer concave face, colored by its electrostatic potential (red: −52 mV, blue: 52 mV), calculated at pH 7 and 150 mM NaCl with APBS . Two cavities are marked with yellow open stars. d) A detail view of monomer B showing the salt bridge network formed by residues Glu29/Asp33/Lys260 as well as the disulphide bond established by cysteine residues 250 and 263. Amino acid residues are labelled in one-letter code; the secondary structure elements to which they belong are also labelled.
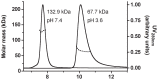
Figure 2. Oligomeric state of ATV in solution.
About 120  g of purified ATV
g of purified ATV were subjected to size-exclusion chromatography coupled to MALS/RI/UV detectors as described in the Experimental section. Two chromatograms/mass analyses are combined in this figure, showing the elution of the tetrameric (left) and dimeric (right) forms of ATV
were subjected to size-exclusion chromatography coupled to MALS/RI/UV detectors as described in the Experimental section. Two chromatograms/mass analyses are combined in this figure, showing the elution of the tetrameric (left) and dimeric (right) forms of ATV , obtained at pH 7.4 and 3.6, respectively. The molar mass (dotted lines), derived from refractive index measurements, and the absorption at 280 nm (full line) were plotted as functions of the elution volume around the peaks. The weight-averaged molar mass (M_w_) values determined by the ASTRA software are indicated.
, obtained at pH 7.4 and 3.6, respectively. The molar mass (dotted lines), derived from refractive index measurements, and the absorption at 280 nm (full line) were plotted as functions of the elution volume around the peaks. The weight-averaged molar mass (M_w_) values determined by the ASTRA software are indicated.
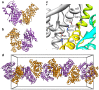
Figure 3. Crystal packing of ATV
 . a) Overall view of the open and b) closed dimers found in the second crystal form. In both cases the pseudo-two-fold axis is vertical in the plane of the paper. c) View of the interface between monomer A (colored as in in Figure 1) and monomer B (grey) in the closed dimer. Amino acid residues involved in cross-monomer salt bridges are labelled in one-letter code. The secondary structure elements that support them are also labelled. d) A view of the dimers arranged as a continuous helical fiber in the crystal. The edges of a unit cell box are shown in grey color.
. a) Overall view of the open and b) closed dimers found in the second crystal form. In both cases the pseudo-two-fold axis is vertical in the plane of the paper. c) View of the interface between monomer A (colored as in in Figure 1) and monomer B (grey) in the closed dimer. Amino acid residues involved in cross-monomer salt bridges are labelled in one-letter code. The secondary structure elements that support them are also labelled. d) A view of the dimers arranged as a continuous helical fiber in the crystal. The edges of a unit cell box are shown in grey color.
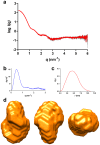
Figure 4. SAXS analysis of ATV
 . a) SAXS intensity as a function of the momentum transfer. This profile corresponds to the measurements taken at 4.7 mg/ml protein cocentration and pH 8.5. Average values are in red and the standard error in grey. b) The Kratky plot (see text for details) corresponds to a folded protein. c) Pair-distance distribution, P(r), function of the data shown in panel a). d) Three orthogonal views of the ab initio envelope calculated imposing orthorhombic symmetry.
. a) SAXS intensity as a function of the momentum transfer. This profile corresponds to the measurements taken at 4.7 mg/ml protein cocentration and pH 8.5. Average values are in red and the standard error in grey. b) The Kratky plot (see text for details) corresponds to a folded protein. c) Pair-distance distribution, P(r), function of the data shown in panel a). d) Three orthogonal views of the ab initio envelope calculated imposing orthorhombic symmetry.
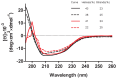
Figure 5. Circular dichroism spectra of ATV
 . Mean residue ellipticity spectra recorded at pH 7.2 (black lines) and pH 0 (red lines), either at 20°C (full lines) or at 80°C (dashed lines). The content in helices and strands, as determined by deconvolution of the spectra, is shown (see text for details).
. Mean residue ellipticity spectra recorded at pH 7.2 (black lines) and pH 0 (red lines), either at 20°C (full lines) or at 80°C (dashed lines). The content in helices and strands, as determined by deconvolution of the spectra, is shown (see text for details).
Similar articles
- The thermo- and acido-stable ORF-99 from the archaeal virus AFV1.
Goulet A, Spinelli S, Blangy S, van Tilbeurgh H, Leulliot N, Basta T, Prangishvili D, Cambillau C, Campanacci V. Goulet A, et al. Protein Sci. 2009 Jun;18(6):1316-20. doi: 10.1002/pro.122. Protein Sci. 2009. PMID: 19472363 Free PMC article. - Chaperone role for proteins p618 and p892 in the extracellular tail development of Acidianus two-tailed virus.
Scheele U, Erdmann S, Ungewickell EJ, Felisberto-Rodrigues C, Ortiz-Lombardía M, Garrett RA. Scheele U, et al. J Virol. 2011 May;85(10):4812-21. doi: 10.1128/JVI.00072-11. Epub 2011 Mar 2. J Virol. 2011. PMID: 21367903 Free PMC article. - Getting the best out of long-wavelength X-rays: de novo chlorine/sulfur SAD phasing of a structural protein from ATV.
Goulet A, Vestergaard G, Felisberto-Rodrigues C, Campanacci V, Garrett RA, Cambillau C, Ortiz-Lombardía M. Goulet A, et al. Acta Crystallogr D Biol Crystallogr. 2010 Mar;66(Pt 3):304-8. doi: 10.1107/S0907444909051798. Epub 2010 Feb 12. Acta Crystallogr D Biol Crystallogr. 2010. PMID: 20179342 - Determination of protein oligomeric structure from small-angle X-ray scattering.
Korasick DA, Tanner JJ. Korasick DA, et al. Protein Sci. 2018 Apr;27(4):814-824. doi: 10.1002/pro.3376. Epub 2018 Feb 10. Protein Sci. 2018. PMID: 29352739 Free PMC article. Review. - Structural characterization of proteins and complexes using small-angle X-ray solution scattering.
Mertens HD, Svergun DI. Mertens HD, et al. J Struct Biol. 2010 Oct;172(1):128-41. doi: 10.1016/j.jsb.2010.06.012. Epub 2010 Jun 15. J Struct Biol. 2010. PMID: 20558299 Review.
Cited by
- Unification of the globally distributed spindle-shaped viruses of the Archaea.
Krupovic M, Quemin ER, Bamford DH, Forterre P, Prangishvili D. Krupovic M, et al. J Virol. 2014 Feb;88(4):2354-8. doi: 10.1128/JVI.02941-13. Epub 2013 Dec 11. J Virol. 2014. PMID: 24335300 Free PMC article. - Archaeal viruses, not archaeal phages: an archaeological dig.
Abedon ST, Murray KL. Abedon ST, et al. Archaea. 2013;2013:251245. doi: 10.1155/2013/251245. Epub 2013 Apr 7. Archaea. 2013. PMID: 23653528 Free PMC article. Review. - Viruses of archaea: Structural, functional, environmental and evolutionary genomics.
Krupovic M, Cvirkaite-Krupovic V, Iranzo J, Prangishvili D, Koonin EV. Krupovic M, et al. Virus Res. 2018 Jan 15;244:181-193. doi: 10.1016/j.virusres.2017.11.025. Epub 2017 Nov 22. Virus Res. 2018. PMID: 29175107 Free PMC article. Review. - A survey of protein structures from archaeal viruses.
Dellas N, Lawrence CM, Young MJ. Dellas N, et al. Life (Basel). 2013 Jan 24;3(1):118-30. doi: 10.3390/life3010118. Life (Basel). 2013. PMID: 25371334 Free PMC article. Review.
References
- Bergh O, Borsheim KY, Bratbak G, Heldal M (1989) High abundance of viruses found in aquatic environments. Nature 340: 467–468. - PubMed
- Suttle CA (2007) Marine viruses–major players in the global ecosystem. Nat Rev Microbiol 5: 801–812. - PubMed
- Torsvik T, Dundas ID (1974) Bacteriophage of Halobacterium salinarium. Nature 248: 680–681. - PubMed
- Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, et al. (1994) Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst Appl Microbiol 16: 609–628.
Publication types
MeSH terms
Substances
Grants and funding
C.F.-R. was funded by a doctoral fellowship from the Fundação para a Ciência e a Tecnologia (http://www.fct.pt/), Portugal (SFRH/BD/44380/2008). The Ville de Marseille (http://www.marseille.fr) provided a financial installation aid to M.O.-L. The Archaea Centre, Copenhagen was supported by a grant from the Danish Natural Science Research Council. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Research Materials