Predicting the functional effect of amino acid substitutions and indels - PubMed (original) (raw)
Predicting the functional effect of amino acid substitutions and indels
Yongwook Choi et al. PLoS One. 2012.
Abstract
As next-generation sequencing projects generate massive genome-wide sequence variation data, bioinformatics tools are being developed to provide computational predictions on the functional effects of sequence variations and narrow down the search of casual variants for disease phenotypes. Different classes of sequence variations at the nucleotide level are involved in human diseases, including substitutions, insertions, deletions, frameshifts, and non-sense mutations. Frameshifts and non-sense mutations are likely to cause a negative effect on protein function. Existing prediction tools primarily focus on studying the deleterious effects of single amino acid substitutions through examining amino acid conservation at the position of interest among related sequences, an approach that is not directly applicable to insertions or deletions. Here, we introduce a versatile alignment-based score as a new metric to predict the damaging effects of variations not limited to single amino acid substitutions but also in-frame insertions, deletions, and multiple amino acid substitutions. This alignment-based score measures the change in sequence similarity of a query sequence to a protein sequence homolog before and after the introduction of an amino acid variation to the query sequence. Our results showed that the scoring scheme performs well in separating disease-associated variants (n = 21,662) from common polymorphisms (n = 37,022) for UniProt human protein variations, and also in separating deleterious variants (n = 15,179) from neutral variants (n = 17,891) for UniProt non-human protein variations. In our approach, the area under the receiver operating characteristic curve (AUC) for the human and non-human protein variation datasets is ∼0.85. We also observed that the alignment-based score correlates with the deleteriousness of a sequence variation. In summary, we have developed a new algorithm, PROVEAN (Protein Variation Effect Analyzer), which provides a generalized approach to predict the functional effects of protein sequence variations including single or multiple amino acid substitutions, and in-frame insertions and deletions. The PROVEAN tool is available online at http://provean.jcvi.org.
Conflict of interest statement
Competing Interests: The authors have the following competing interests: The authors have developed a new algorithm, PROVEAN (Protein Variation Effect Analyzer), which provides a generalized approach to predict the functional effects of protein sequence variations including single or multiple amino acid substitutions, and in-frame insertions and deletions. The PROVEAN tool is available online at http://provean.jcvi.org. There are no further patents, products in development or marketed products to declare. This does not alter the authors' adherence to all the PLOS ONE policies on sharing data and materials, as detailed online in the guide for authors.
Figures
Figure 1. Examples of computing and interpreting delta alignment scores for six different known variations, (A) deleterious substitution (MIM:151623), (B) neutral substitution (dbSNP:rs1042522), (C) deleterious deletion (MIM:219700), (D) neutral deletion (dbSNP:rs72471101), (E) deleterious insertion (MIM:164200), and (F) neutral insertion (dbSNP:rs10625857) with respect to the selected homologous proteins.
The amino acid residue replaced, deleted, or inserted is indicated by an arrow, and the difference between two alignments is indicated by a rectangle. Low delta scores are interpreted as deleterious, and high delta scores are interpreted as neutral. The BLOSUM62 and gap penalties of 10 for opening and 1 for extension were used.
Figure 2. An overview of the PROVEAN procedure.
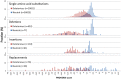
Figure 3. PROVEAN score distribution for deleterious and neutral human protein variations.
For all classes of variations including substitutions, indels, and replacements, the distribution shows a distinct separation between the deleterious and neutral variations.
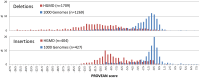
Figure 4. PROVEAN score distribution of deletions and insertions collected from the Human Gene Mutation Database (HGMD) and the 1000 Genomes Project.
Only mutations annotated as “disease-causing” were collected from the HGMD. The distribution shows a distinct separation between the two datasets.
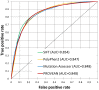
Figure 5. ROC curves of four different prediction tools for single amino acid substitutions found in human and non-human proteins. All tools show a similar predictive ability with the AUC value of ∼0.85.
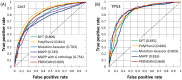
Figure 6. ROC curves of different prediction tools for single amino acid substitutions in (A) E. coli lac repressor protein and (B) human TP53 tumor suppressor protein.
The AUC values are shown in the legend. The top two performers for LacI were PolyPhen-2 and PROVEAN, and those for TP53 were Mutation Assessor and PROVEAN.
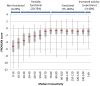
Figure 7. PROVEAN score distribution of TP53 variations binned into 15 classes based on transactivation levels.
For each class, a box plot is shown. The vertical line shows the whole range of delta scores, the thick horizontal line shows the median, and the gray rectangle shows the interquartile range (25%–75%). The PROVEAN score increases and correlates with median transactivation level for the non-functional and partially functional classes of variations.
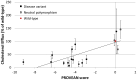
Figure 8. Correlation of cholesterol efflux values with the PROVEAN scores for ABCA1 variations.
In general, an increase in score (i.e. less deleterious effects) correlates with an increase in cholesterol efflux activity.
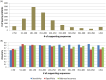
Figure 9. Number of supporting sequences used for the Uniprot human proteins carrying neutral or deleterious variants.
(A) Distribution of the 11,990 human proteins based on the number of supporting sequences used for PROVEAN prediction. (B) Prediction accuracy achieved with respect to the number of supporting sequences. The observed accuracy is consistently above 73%, except in cases when the number of supporting sequences drops below 50.
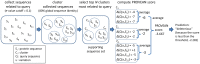
Figure 10. Computing the PROVEAN score.
For simplicity, only the top three clusters were included in building the supporting sequence set in this example.
Similar articles
- PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels.
Choi Y, Chan AP. Choi Y, et al. Bioinformatics. 2015 Aug 15;31(16):2745-7. doi: 10.1093/bioinformatics/btv195. Epub 2015 Apr 6. Bioinformatics. 2015. PMID: 25851949 Free PMC article. - SIFT Indel: predictions for the functional effects of amino acid insertions/deletions in proteins.
Hu J, Ng PC. Hu J, et al. PLoS One. 2013 Oct 23;8(10):e77940. doi: 10.1371/journal.pone.0077940. eCollection 2013. PLoS One. 2013. PMID: 24194902 Free PMC article. - Quantitative prediction of the effect of genetic variation using hidden Markov models.
Liu M, Watson LT, Zhang L. Liu M, et al. BMC Bioinformatics. 2014 Jan 9;15:5. doi: 10.1186/1471-2105-15-5. BMC Bioinformatics. 2014. PMID: 24405700 Free PMC article. - An integrative approach to predicting the functional effects of small indels in non-coding regions of the human genome.
Ferlaino M, Rogers MF, Shihab HA, Mort M, Cooper DN, Gaunt TR, Campbell C. Ferlaino M, et al. BMC Bioinformatics. 2017 Oct 6;18(1):442. doi: 10.1186/s12859-017-1862-y. BMC Bioinformatics. 2017. PMID: 28985712 Free PMC article. - Insertions and Deletions: Computational Methods, Evolutionary Dynamics, and Biological Applications.
Redelings BD, Holmes I, Lunter G, Pupko T, Anisimova M. Redelings BD, et al. Mol Biol Evol. 2024 Sep 4;41(9):msae177. doi: 10.1093/molbev/msae177. Mol Biol Evol. 2024. PMID: 39172750 Free PMC article. Review.
Cited by
- Unraveling the protective genetic architecture of COVID-19 in the Brazilian Amazon.
Barros MC, de Souza JES, Gomes DHF, Pinho CT, Silva CS, Braga-da-Silva C, Cavalcante GC, Magalhães L, Azevedo-Pinheiro J, Quaresma JAS, Falcão LFM, Costa PF, Salgado CG, Carneiro TX, Burbano RR, Dos Santos Vieira JR, Santos S, Soares-Souza GB, de Souza SJ, Ribeiro-Dos-Santos Â. Barros MC, et al. Sci Rep. 2024 Nov 9;14(1):27332. doi: 10.1038/s41598-024-78170-3. Sci Rep. 2024. PMID: 39521879 Free PMC article. - Genetic and functional analyses of CTBP2 in anorexia nervosa and body weight regulation.
Giuranna J, Zheng Y, Brandt M, Jall S, Mukherjee A, Shankhwar S, Renner S, Kurapati NK, May C, Peters T, Herpertz-Dahlmann B, Seitz J, de Zwaan M, Herzog W, Ehrlich S, Zipfel S, Giel K, Egberts K, Burghardt R, Föcker M, Marcus K, Keyvani K, Müller TD, Schmitz F, Rajcsanyi LS, Hinney A. Giuranna J, et al. Mol Psychiatry. 2024 Nov 7. doi: 10.1038/s41380-024-02791-3. Online ahead of print. Mol Psychiatry. 2024. PMID: 39511451 - Cross-ancestry analysis identifies genes associated with obesity risk and protection.
Banerjee D, Girirajan S. Banerjee D, et al. medRxiv [Preprint]. 2024 Oct 16:2024.10.13.24315422. doi: 10.1101/2024.10.13.24315422. medRxiv. 2024. PMID: 39484254 Free PMC article. Preprint. - Cryo-EM structure of the human subcortical maternal complex and the associated discovery of infertility-associated variants.
Chi P, Ou G, Liu S, Ma Q, Lu Y, Li J, Li J, Qi Q, Han Z, Zhang Z, Liu Q, Guo L, Chen J, Wang X, Huang W, Li L, Deng D. Chi P, et al. Nat Struct Mol Biol. 2024 Nov;31(11):1798-1807. doi: 10.1038/s41594-024-01396-2. Epub 2024 Oct 8. Nat Struct Mol Biol. 2024. PMID: 39379527 - Determination of the frequency and distribution of APC, PIK3CA, and SMAD4 gene mutations in Ugandan patients with colorectal cancer.
Wismayer R, Matthews R, Whalley C, Kiwanuka J, Kakembo FE, Thorn S, Wabinga H, Odida M, Tomlinson I. Wismayer R, et al. BMC Cancer. 2024 Sep 30;24(1):1212. doi: 10.1186/s12885-024-12967-3. BMC Cancer. 2024. PMID: 39350061 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials