SMN is required for sensory-motor circuit function in Drosophila - PubMed (original) (raw)
SMN is required for sensory-motor circuit function in Drosophila
Wendy L Imlach et al. Cell. 2012.
Abstract
Spinal muscular atrophy (SMA) is a lethal human disease characterized by motor neuron dysfunction and muscle deterioration due to depletion of the ubiquitous survival motor neuron (SMN) protein. Drosophila SMN mutants have reduced muscle size and defective locomotion, motor rhythm, and motor neuron neurotransmission. Unexpectedly, restoration of SMN in either muscles or motor neurons did not alter these phenotypes. Instead, SMN must be expressed in proprioceptive neurons and interneurons in the motor circuit to nonautonomously correct defects in motor neurons and muscles. SMN depletion disrupts the motor system subsequent to circuit development and can be mimicked by the inhibition of motor network function. Furthermore, increasing motor circuit excitability by genetic or pharmacological inhibition of K(+) channels can correct SMN-dependent phenotypes. These results establish sensory-motor circuit dysfunction as the origin of motor system deficits in this SMA model and suggest that enhancement of motor neural network activity could ameliorate the disease.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
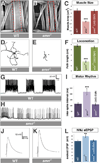
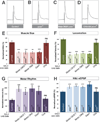
Figure 1. smn mutants have reduced muscle size, decreased locomotion, defective motor rhythm and aberrant NMJ neurotransmitter release
A–C. Sample images of muscles from segment A3 of control (A) and _smn_X7 mutant (B) third instar larvae labeled with TRITC-phallodin show a reduction muscle surface area (C) that is fully rescued by ubiquitous expression of UAS-flag-SMN driven by Da-Gal4 (genotype: Da-Gal4/UAS::flagSMN; _smn_X7/_smn_X7). D–F. 10 sample superimposed 60 second larval locomotion path traces from control (D) and _smn_X7 mutants (E). smn mutant larvae have reduced velocity compared to controls that corrected by ubiquitous expression of transgenic SMN (F). G–I. Recordings from muscle 6 in segment A1 of semi-intact larval preparations where the brain, ventral nerve cord and motor neurons are intact. Control larva produce a regular motor rhythm with periodic bursting activity corresponding to peristaltic muscle contractions (G). smn mutant larvae have an irregular motor pattern with short and uncoordinated bursts as shown by an increase in the average inter-spike interval (I) that is rescued by ubiquitous expression of SMN. J–L. Representative traces recorded from muscle 6 of segment A3 in control (J) and smnX7 mutant (K) larva. _smn_X7 mutants have increased evoked Excitatory Post-Synaptic Potential (eEPSP) amplitude than controls (K). This is corrected by ubiquitous expression of SMN (L). Data are represented as mean +/− SEM, **=p<0.01, ***=p<0.001. See also Figure S1 and Table S1.
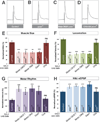
Figure 2. SMN expression is required in neurons but not muscles to rescue smn mutants
A–D. Sample images of muscles from segment A3 of (A) control, (B) _smn_X7 mutant, (C) _smn_X7 mutants with transgenic SMN expression only in muscles (G14-Gal4/UAS::flagSMN; _smn_X7/_smn_X7) or (D) neurons (nsyb-Gal4/UAS::flagSMN; _smn_X7/_smn_X7). Restoration of SMN expression in muscles has no effect on muscle size however restoration in neurons fully rescues muscle surface area. E–H. Quantification of muscle surface area (E), locomotion (F), motor rhythm (G) and NMJ eEPSP amplitude (H) normalized to controls. Expression of transgenic SMN in neurons rescues all of smn mutant phenotypes while expression in muscles does not. Data are represented as mean +/− SEM, **=p<0.01, ***=p<0.001. See also Figure S2.
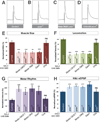
Figure 3. SMN expression is required in cholinergic neurons and not motor neurons
A–D. Representative traces of control (A), _smn_X7 mutant (B), transgenic SMN expressed in the motor neurons of smn mutants (OK371-Gal4/UAS::SMN; _smn_X7/_smn_X7) (C), transgenic SMN expressed in the cholinergic neurons of smn mutants (Cha-Gal4/UAS::SMN; _smn_X7/_smn_X7) (D). Expression of transgenic SMN in motor neurons does not restore NMJ eEPSP amplitudes in smn mutants however expression of SMN on cholinergic neurons does. E–F. Quantification of muscle surface area (E), locomotion (F), motor rhythm (G) and NMJ eEPSP amplitude (H) normalized to controls. Expression of transgenic SMN in the motor neurons of smn mutants with OK371-Gal4 or OK6-Gal4 or in GABAnergic neurons with GAD1-Gal4 does not rescue any phenotype. Expression of transgenic SMN in cholinergic neurons rescues all phenotypes (D). Data are represented as mean +/− SEM, *= p<0.05, **=p<0.01, ***=p<0.001. Significance was calculated versus controls except where otherwise indicated.
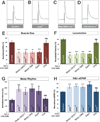
Figure 4. SMN is required in both proprioceptive and central cholinergic neurons
A. Expression of pattern of cholinergic neuron Gal4 lines (dark green). Cha-Gal4 is expressed in both central and sensory cholinergic neurons. Clh201-Gal4 is only expressed in md and es sensory neurons. 1003.3-Gal4, ppk-Gal4 and NP2225-Gal4 are expressed in subsets of md, es or ch sensory neurons. Bright green indicates the ability to rescue of smn mutant phenotypes. B,C. UAS::CD8-GFP labeling the axons of bd and type I md sensory neurons with NP2225-Gal4 in the ventral nerve cord of wild-type (B) or _smn_X7 mutants (C). Sensory axons project normally into the CNS in smn mutants. D–G. Quantification of muscle surface area (D), locomotion (E), motor rhythm (F) and NMJ eEPSP amplitude (G) normalized to controls (genotype: Gal4/UAS::flagSMN; _smn_X7/_smn_X7). Expression of transgenic SMN in both central and sensory cholinergic neurons in smn mutants with Cha-Gal4 fully rescues all phenotypes. Restoration of SMN all sensory neurons or only proprioceptive type I md and bd neurons with NP2225-Gal4 increases muscle size and fully rescues motor rhythm and NMJ eEPSP amplitude but does not rescue locomotion. In contrast, restoration of SMN in with 1003.3-Gal4 or ppk-Gal4 does not rescue any smn mutant phenotype. Scale bar = 10µm. Data are represented as mean +/− SEM, *=p<0.05, **=p<0.01, ***=p<0.001. Significance was calculated versus controls except where otherwise indicated.
Figure 5. Restoration of SMN after embryogenesis rescues smn mutants
A. Schematic of transgenic SMN induction in the nervous system. RU486 is required for the activation of transgene induction by geneswitch Gal4. Elav::geneswitch/UAS::flagSMN; _smn_X7/_smn_X7 larva were transferred to either vehicle media or RU486 containing media immediately, 48 hours or 96 hours after hatching. B. Representative traces recorded from smn mutants that were cultured on either vehicle media or RU486 media 0, 48 or 96 hours after hatching. Induction of SMN at every each time-point fully restored normal eEPSP amplitude. C–F. Quantification of muscle surface area (C), locomotion (D), motor rhythm (E) and NMJ eEPSP amplitude (F) normalized to controls. Muscle size, locomotion and motor rhythm is fully rescued if transgenic SMN is induced immediately after hatching, but if SMN induction is delayed rescue is incomplete. Induction of SMN for only 48 hours is however sufficient to completely restore normal NMJ eEPSP amplitude. Data are represented as mean +/− SEM, *=p<0.05, **=p<0.01, ***=p<0.001. Significance was calculated versus controls except where otherwise indicated.
Figure 6. Inhibiting cholinergic neuron activity mimics smn mutant phenotypes
A. Representative traces recorded from the NMJ of control or UAS-human Kir2.1 or UASPLTXII expressed in cholinergic neurons with Cha-Gal4. Inhibiting cholinergic neuron excitability with Kir2.1 or neurotransmitter release with PLTXII increases motor neuron NMJ eEPSP amplitude. B. Expression of Kir2.1 or PLTX in cholinergic neurons disrupts rhythmic motor activity. C–F. Quantification of muscle surface area (C), locomotion (D), motor rhythm (E) and NMJ eEPSP amplitude (F) normalized to controls. Expression of Kir2.1 or PLTXII in cholinergic neurons does not alter muscle size but does reduce locomotor speed, disrupt motor rhythm and increases NMJ eEPSP amplitude. Data are represented as mean +/− SEM, *=p<0.05, ***=p<0.001.
Figure 7. Genetic or pharmacological inhibition of K+ channels ameliorates smn mutant phenotypes
A–C. Locomotion path traces from (A) control, (B) smn mutants and (C) smn mutants expressing a UAS dominant negative Shaker K+ channel (UAS-SDN) in cholinergic neurons with Cha-Gal4. Expressing SDN increases rescues the locomotion of smn mutants. D–G. Quantification of muscle surface area (D), locomotion (E), motor rhythm (F) and NMJ eEPSP amplitude (G) normalized to controls. Expression of SDN in cholinergic neurons with Cha-Gal4 restores muscle size (D), locomotion (E), motor rhythm (F) and NMJ eEPSP (G) of smn mutants to control levels. Addition of 2mM 4-Aminopyridine (4-AP) to culture media throughout larval development does not alter muscle size in control animals but increases the muscle size of smn mutants (D). 4-AP administration inhibits locomotion, motor rhythm and eEPSP size in control animals. Administration of 4-AP to smn mutants corrects locomotion (E) and NMJ eEPSP (G) to levels not significantly different from control 4-AP treated animals and substantially corrects defects in motor rhythm (F). Data are represented as mean +/− SEM, *=p<0.05, **=p<0.01, ***=p<0.001. Significance was calculated versus controls except where otherwise indicated.
Comment in
- A circuit mechanism for neurodegeneration.
Roselli F, Caroni P. Roselli F, et al. Cell. 2012 Oct 12;151(2):250-2. doi: 10.1016/j.cell.2012.09.030. Cell. 2012. PMID: 23063119
Similar articles
- Modeling spinal muscular atrophy in Drosophila links Smn to FGF signaling.
Sen A, Yokokura T, Kankel MW, Dimlich DN, Manent J, Sanyal S, Artavanis-Tsakonas S. Sen A, et al. J Cell Biol. 2011 Feb 7;192(3):481-95. doi: 10.1083/jcb.201004016. J Cell Biol. 2011. PMID: 21300852 Free PMC article. - An SMN-dependent U12 splicing event essential for motor circuit function.
Lotti F, Imlach WL, Saieva L, Beck ES, Hao le T, Li DK, Jiao W, Mentis GZ, Beattie CE, McCabe BD, Pellizzoni L. Lotti F, et al. Cell. 2012 Oct 12;151(2):440-54. doi: 10.1016/j.cell.2012.09.012. Cell. 2012. PMID: 23063131 Free PMC article. - WDR79/TCAB1 plays a conserved role in the control of locomotion and ameliorates phenotypic defects in SMA models.
Di Giorgio ML, Esposito A, Maccallini P, Micheli E, Bavasso F, Gallotta I, Vernì F, Feiguin F, Cacchione S, McCabe BD, Di Schiavi E, Raffa GD. Di Giorgio ML, et al. Neurobiol Dis. 2017 Sep;105:42-50. doi: 10.1016/j.nbd.2017.05.005. Epub 2017 May 11. Neurobiol Dis. 2017. PMID: 28502804 - Fishing for a mechanism: using zebrafish to understand spinal muscular atrophy.
Beattie CE, Carrel TL, McWhorter ML. Beattie CE, et al. J Child Neurol. 2007 Aug;22(8):995-1003. doi: 10.1177/0883073807305671. J Child Neurol. 2007. PMID: 17761655 Review. - Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease.
Monani UR. Monani UR. Neuron. 2005 Dec 22;48(6):885-96. doi: 10.1016/j.neuron.2005.12.001. Neuron. 2005. PMID: 16364894 Review.
Cited by
- Clinical and Genetic Profiles of 5q- and Non-5q-Spinal Muscular Atrophy Diseases in Pediatric Patients.
Nishio H, Niba ETE, Saito T, Okamoto K, Lee T, Takeshima Y, Awano H, Lai PS. Nishio H, et al. Genes (Basel). 2024 Sep 30;15(10):1294. doi: 10.3390/genes15101294. Genes (Basel). 2024. PMID: 39457418 Free PMC article. Review. - Motor fiber function in spinal muscular atrophy-analysis of conduction velocity distribution.
Koszewicz M, Ubysz J, Dziadkowiak E, Wieczorek M, Budrewicz S. Koszewicz M, et al. Front Neurol. 2023 Dec 7;14:1305497. doi: 10.3389/fneur.2023.1305497. eCollection 2023. Front Neurol. 2023. PMID: 38192575 Free PMC article. - Connectivity and circuitry in a dish versus in a brain.
Chinchalongporn V, Koppensteiner P, Prè D, Thangnipon W, Bilo L, Arancio O. Chinchalongporn V, et al. Alzheimers Res Ther. 2015 Jun 4;7(1):44. doi: 10.1186/s13195-015-0129-y. eCollection 2015. Alzheimers Res Ther. 2015. PMID: 26045718 Free PMC article. - Impact of Aging on Proprioceptive Sensory Neurons and Intrafusal Muscle Fibers in Mice.
Vaughan SK, Stanley OL, Valdez G. Vaughan SK, et al. J Gerontol A Biol Sci Med Sci. 2017 Jun 1;72(6):771-779. doi: 10.1093/gerona/glw175. J Gerontol A Biol Sci Med Sci. 2017. PMID: 27688482 Free PMC article. - Therapy development for spinal muscular atrophy: perspectives for muscular dystrophies and neurodegenerative disorders.
Jablonka S, Hennlein L, Sendtner M. Jablonka S, et al. Neurol Res Pract. 2022 Jan 4;4(1):2. doi: 10.1186/s42466-021-00162-9. Neurol Res Pract. 2022. PMID: 34983696 Free PMC article. Review.
References
- Baines RA. Development of motoneuron electrical properties and motor output. Int Rev Neurobiol. 2006;75:91–103. - PubMed
- Cattaert D, Birman S. Blockade of the central generator of locomotor rhythm by noncompetitive NMDA receptor antagonists in Drosophila larvae. J Neurobiol. 2001;48:58–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases