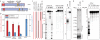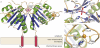The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis - PubMed (original) (raw)
. 2012 Nov 8;491(7423):279-83.
doi: 10.1038/nature11502. Epub 2012 Oct 14.
Affiliations
- PMID: 23064227
- PMCID: PMC3493678
- DOI: 10.1038/nature11502
The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis
Jonathan J Ipsaro et al. Nature. 2012.
Abstract
PIWI-family proteins and their associated small RNAs (piRNAs) act in an evolutionarily conserved innate immune mechanism to provide essential protection for germ-cell genomes against the activity of mobile genetic elements. piRNA populations comprise a molecular definition of transposons, which permits them to distinguish transposons from host genes and selectively silence them. piRNAs can be generated in two distinct ways, forming either primary or secondary piRNAs. Primary piRNAs come from discrete genomic loci, termed piRNA clusters, and seem to be derived from long, single-stranded precursors. The biogenesis of primary piRNAs involves at least two nucleolytic steps. An unknown enzyme cleaves piRNA cluster transcripts to generate monophosphorylated piRNA 5' ends. piRNA 3' ends are probably formed by exonucleolytic trimming, after a piRNA precursor is loaded into its PIWI partner. Secondary piRNAs arise during the adaptive 'ping-pong' cycle, with their 5' termini being formed by the activity of PIWIs themselves. A number of proteins have been implicated genetically in primary piRNA biogenesis. One of these, Drosophila melanogaster Zucchini, is a member of the phospholipase-D family of phosphodiesterases, which includes both phospholipases and nucleases. Here we produced a dimeric, soluble fragment of the mouse Zucchini homologue (mZuc; also known as PLD6) and show that it possesses single-strand-specific nuclease activity. A crystal structure of mZuc at 1.75 Å resolution indicates greater architectural similarity to phospholipase-D family nucleases than to phospholipases. Together, our data suggest that the Zucchini proteins act in primary piRNA biogenesis as nucleases, perhaps generating the 5' ends of primary piRNAs.
Figures
Figure 1. mZuc acts as a nuclease but not a phospholipase in vitro
a) The domain architecture of mZuc and Zuc are similar with an N-terminal transmembrane helix (TM, red) and a cytoplasmic domain, which contains the catalytic HKD motif (gold). The construct used for crystallization is indicated as a dashed box. Residue numbers delineating each domain indicated below each schematic. b) Phosphodiesterase activity for mZuc, its catalytic mutant (H153N), and a known phospholipase (Phospholipase D from Streptomyces chromofuscus, scPLD) were monitored by SRM-MS. Levels of the cardiolipin substrate and the expected phophatidic acid product (PA) are shown for each reaction. Error bars indicate standard deviation (n=3). c) DNase activity of recombinant mZuc was monitored using 50 nt 5'-32P-labeled DNA (red). Single-stranded (ss), double-stranded (ds), and partially double-stranded substrates were incubated with wild-type (wt) or catalytic mutant H153N enzyme (as indicated), and the resulting products separated by urea-PAGE. d) mZuc or the H153N mutant were incubated in the presence of 32PO43- followed by SDS-PAGE and transfer to a nitrocellulose membrane. e) The DNase of mZuc was measured in the presence of Na3VO4. scPLD is shown as control.
Figure 2. mZuc acts as a single-strand specific endoribonuclease in vitro
a) mZuc was incubated with ss, ds, and partially dsRNA substrates in the absence and presence of Na3VO4 as indicated. b) mZuc RNA cleavage products (from reactions shown in panel a, as indicated below) were tested for sensitivity to β-elimination and accessibility for polyadenylation.
Figure 3. Crystal structure of mZuc
a) The overall structure of the mZuc dimer is shown as a ribbon diagram. Helices are in green, strands in blue, and loops in beige. Each monomer binds one Zn2+ (yellow) in an extended zinc wing. The active site histidine residues (His153) are highlighted in red. b) A close-up of the zinc wing consisting of residues Cys49, Cys66, Cys68, and His72 is shown. c) A detailed view of mZuc co-crystallized with tungstate bound in the active site is presented.
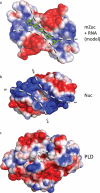
Figure 4. Electrostatic surfaces of PLD family proteins indicate distinct binding surfaces for specific substrates
a) The electrostatic surface for mZuc displays a long, narrow, positively-charged grove laying across the zinc wings and active site. A short RNA molecule was manually built into the structure of mZuc, then subjected to energy minimization using GROMACS. The minimized model shows the phosphates of the RNA backbone positioned in the most positively charged areas of the groove with the bases extending away from the dimer core. b) In Nuc (PDB ID 1BYR), which acts on double-stranded DNA, the equivalent groove is significantly wider. c) A bone fide phospholipase, PLD (PDB ID 2ZE9), uses the same active site architecture in a strikingly different structural context. PLD has a small pocket for binding phospholipids, rather than an elongated groove. Each surface depicts the solvent-accessible surface contoured at ± 2 kB T/e using ABPS.
Similar articles
- Structure and function of Zucchini endoribonuclease in piRNA biogenesis.
Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, Ishitani R, Siomi H, Siomi MC, Nureki O. Nishimasu H, et al. Nature. 2012 Nov 8;491(7423):284-7. doi: 10.1038/nature11509. Epub 2012 Oct 14. Nature. 2012. PMID: 23064230 - Crystal structure of the primary piRNA biogenesis factor Zucchini reveals similarity to the bacterial PLD endonuclease Nuc.
Voigt F, Reuter M, Kasaruho A, Schulz EC, Pillai RS, Barabas O. Voigt F, et al. RNA. 2012 Dec;18(12):2128-34. doi: 10.1261/rna.034967.112. Epub 2012 Oct 19. RNA. 2012. PMID: 23086923 Free PMC article. - Zucchini consensus motifs determine the mechanism of pre-piRNA production.
Izumi N, Shoji K, Suzuki Y, Katsuma S, Tomari Y. Izumi N, et al. Nature. 2020 Feb;578(7794):311-316. doi: 10.1038/s41586-020-1966-9. Epub 2020 Jan 29. Nature. 2020. PMID: 31996847 - The piRNA pathway in Drosophila ovarian germ and somatic cells.
Sato K, Siomi MC. Sato K, et al. Proc Jpn Acad Ser B Phys Biol Sci. 2020;96(1):32-42. doi: 10.2183/pjab.96.003. Proc Jpn Acad Ser B Phys Biol Sci. 2020. PMID: 31932527 Free PMC article. Review. - PIWI-Interacting RNA in Drosophila: Biogenesis, Transposon Regulation, and Beyond.
Yamashiro H, Siomi MC. Yamashiro H, et al. Chem Rev. 2018 Apr 25;118(8):4404-4421. doi: 10.1021/acs.chemrev.7b00393. Epub 2017 Dec 27. Chem Rev. 2018. PMID: 29281264 Review.
Cited by
- Hsp90 facilitates accurate loading of precursor piRNAs into PIWI proteins.
Izumi N, Kawaoka S, Yasuhara S, Suzuki Y, Sugano S, Katsuma S, Tomari Y. Izumi N, et al. RNA. 2013 Jul;19(7):896-901. doi: 10.1261/rna.037200.112. Epub 2013 May 16. RNA. 2013. PMID: 23681506 Free PMC article. - Maelstrom functions in the production of Siwi-piRISC capable of regulating transposons in Bombyx germ cells.
Namba Y, Iwasaki YW, Nishida KM, Nishihara H, Sumiyoshi T, Siomi MC. Namba Y, et al. iScience. 2022 Feb 11;25(3):103914. doi: 10.1016/j.isci.2022.103914. eCollection 2022 Mar 18. iScience. 2022. PMID: 35243263 Free PMC article. - The USTC co-opts an ancient machinery to drive piRNA transcription in C. elegans.
Weng C, Kosalka J, Berkyurek AC, Stempor P, Feng X, Mao H, Zeng C, Li WJ, Yan YH, Dong MQ, Morero NR, Zuliani C, Barabas O, Ahringer J, Guang S, Miska EA. Weng C, et al. Genes Dev. 2019 Jan 1;33(1-2):90-102. doi: 10.1101/gad.319293.118. Epub 2018 Dec 19. Genes Dev. 2019. PMID: 30567997 Free PMC article. - Armitage determines Piwi-piRISC processing from precursor formation and quality control to inter-organelle translocation.
Yamashiro H, Negishi M, Kinoshita T, Ishizu H, Ohtani H, Siomi MC. Yamashiro H, et al. EMBO Rep. 2020 Feb 5;21(2):e48769. doi: 10.15252/embr.201948769. Epub 2019 Dec 12. EMBO Rep. 2020. PMID: 31833223 Free PMC article. - Insights in piRNA targeting rules.
van Wolfswinkel JC. van Wolfswinkel JC. Wiley Interdiscip Rev RNA. 2023 Aug 26:e1811. doi: 10.1002/wrna.1811. Online ahead of print. Wiley Interdiscip Rev RNA. 2023. PMID: 37632327
References
- Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi:10.1016/j.cell.2007.01.043. - PubMed
- Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3' end formation of PIWI-interacting RNAs in vitro. Mol Cell. 2011;43:1015–1022. doi:10.1016/j.molcel.2011.07.029. - PubMed
- Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi:10.1126/science.1140494. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM097888/GM/NIGMS NIH HHS/United States
- R01GM062534/GM/NIGMS NIH HHS/United States
- F32GM97888/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM062534/GM/NIGMS NIH HHS/United States
- P30 CA045508/CA/NCI NIH HHS/United States
- CA045508/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases