Common genetic variants, acting additively, are a major source of risk for autism - PubMed (original) (raw)
doi: 10.1186/2040-2392-3-9.
Stephan J Sanders, Michael T Murtha, Vanessa Hus, Jennifer K Lowe, A Jeremy Willsey, Daniel Moreno-De-Luca, Timothy W Yu, Eric Fombonne, Daniel Geschwind, Dorothy E Grice, David H Ledbetter, Catherine Lord, Shrikant M Mane, Christa Lese Martin, Donna M Martin, Eric M Morrow, Christopher A Walsh, Nadine M Melhem, Pauline Chaste, James S Sutcliffe, Matthew W State, Edwin H Cook Jr, Kathryn Roeder, Bernie Devlin
Affiliations
- PMID: 23067556
- PMCID: PMC3579743
- DOI: 10.1186/2040-2392-3-9
Common genetic variants, acting additively, are a major source of risk for autism
Lambertus Klei et al. Mol Autism. 2012.
Abstract
Background: Autism spectrum disorders (ASD) are early onset neurodevelopmental syndromes typified by impairments in reciprocal social interaction and communication, accompanied by restricted and repetitive behaviors. While rare and especially de novo genetic variation are known to affect liability, whether common genetic polymorphism plays a substantial role is an open question and the relative contribution of genes and environment is contentious. It is probable that the relative contributions of rare and common variation, as well as environment, differs between ASD families having only a single affected individual (simplex) versus multiplex families who have two or more affected individuals.
Methods: By using quantitative genetics techniques and the contrast of ASD subjects to controls, we estimate what portion of liability can be explained by additive genetic effects, known as narrow-sense heritability. We evaluate relatives of ASD subjects using the same methods to evaluate the assumptions of the additive model and partition families by simplex/multiplex status to determine how heritability changes with status.
Results: By analyzing common variation throughout the genome, we show that common genetic polymorphism exerts substantial additive genetic effects on ASD liability and that simplex/multiplex family status has an impact on the identified composition of that risk. As a fraction of the total variation in liability, the estimated narrow-sense heritability exceeds 60% for ASD individuals from multiplex families and is approximately 40% for simplex families. By analyzing parents, unaffected siblings and alleles not transmitted from parents to their affected children, we conclude that the data for simplex ASD families follow the expectation for additive models closely. The data from multiplex families deviate somewhat from an additive model, possibly due to parental assortative mating.
Conclusions: Our results, when viewed in the context of results from genome-wide association studies, demonstrate that a myriad of common variants of very small effect impacts ASD liability.
Figures
Figure 1
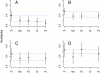
Estimated heritability for Autism Spectrum Disorders from ASD probands (Pr), as well as for their mothers (Mo), fathers (Fa), siblings (Si) and pseudo-controls (Pc). Blue dotted reference line is set to the estimated heritability from probands; the black line marks the expected heritability for first degree relatives; and the gray line marks the expected heritability from pseudo-controls. Expected values derived from simulations mimicking the recruitment strategy producing the samples for (A)-(D). (A) Simons Simplex Collection or SSC data; (B) Autism Genome Project or AGP data; (C) AGP data, only simplex families; (D) AGP data, only multiplex families.
Figure 2
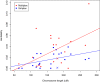
Estimated heritability per chromosome for simplex and muliplex families. In this figure chromosome X is marked distinctly, but each chromosome is mapped by its length.
Similar articles
- How rare and common risk variation jointly affect liability for autism spectrum disorder.
Klei L, McClain LL, Mahjani B, Panayidou K, De Rubeis S, Grahnat AS, Karlsson G, Lu Y, Melhem N, Xu X, Reichenberg A, Sandin S, Hultman CM, Buxbaum JD, Roeder K, Devlin B. Klei L, et al. Mol Autism. 2021 Oct 6;12(1):66. doi: 10.1186/s13229-021-00466-2. Mol Autism. 2021. PMID: 34615521 Free PMC article. - The genetics of autism.
Muhle R, Trentacoste SV, Rapin I. Muhle R, et al. Pediatrics. 2004 May;113(5):e472-86. doi: 10.1542/peds.113.5.e472. Pediatrics. 2004. PMID: 15121991 Review. - The contributions of rare inherited and polygenic risk to ASD in multiplex families.
Cirnigliaro M, Chang TS, Arteaga SA, Pérez-Cano L, Ruzzo EK, Gordon A, Bicks LK, Jung JY, Lowe JK, Wall DP, Geschwind DH. Cirnigliaro M, et al. Proc Natl Acad Sci U S A. 2023 Aug;120(31):e2215632120. doi: 10.1073/pnas.2215632120. Epub 2023 Jul 28. Proc Natl Acad Sci U S A. 2023. PMID: 37506195 Free PMC article. - Cellular and molecular characterization of multiplex autism in human induced pluripotent stem cell-derived neurons.
Lewis EMA, Meganathan K, Baldridge D, Gontarz P, Zhang B, Bonni A, Constantino JN, Kroll KL. Lewis EMA, et al. Mol Autism. 2019 Dec 30;10:51. doi: 10.1186/s13229-019-0306-0. eCollection 2019. Mol Autism. 2019. PMID: 31893020 Free PMC article. - The Yin and Yang of Autism Genetics: How Rare De Novo and Common Variations Affect Liability.
Chaste P, Roeder K, Devlin B. Chaste P, et al. Annu Rev Genomics Hum Genet. 2017 Aug 31;18:167-187. doi: 10.1146/annurev-genom-083115-022647. Epub 2017 Apr 19. Annu Rev Genomics Hum Genet. 2017. PMID: 28426285 Review.
Cited by
- Longitudinal trends and correlation between autism spectrum disorder prevalence and sperm quality parameters (2000-2024): a comprehensive statistical analysis.
Al-Salihy AAS. Al-Salihy AAS. Front Reprod Health. 2024 Aug 22;6:1438049. doi: 10.3389/frph.2024.1438049. eCollection 2024. Front Reprod Health. 2024. PMID: 39239154 Free PMC article. - The Importance of Large-Scale Genomic Studies to Unravel Genetic Risk Factors for Autism.
Nóbrega IS, Teles E Silva AL, Yokota-Moreno BY, Sertié AL. Nóbrega IS, et al. Int J Mol Sci. 2024 May 27;25(11):5816. doi: 10.3390/ijms25115816. Int J Mol Sci. 2024. PMID: 38892002 Free PMC article. Review. - Identification of moderate effect size genes in autism spectrum disorder through a novel gene pairing approach.
Caballero M, Satterstrom FK, Buxbaum JD, Mahjani B. Caballero M, et al. medRxiv [Preprint]. 2024 Apr 4:2024.04.03.24305278. doi: 10.1101/2024.04.03.24305278. medRxiv. 2024. PMID: 38633780 Free PMC article. Preprint. - Structural models of genome-wide covariance identify multiple common dimensions in autism.
de Hoyos L, Barendse MT, Schlag F, van Donkelaar MMJ, Verhoef E, Shapland CY, Klassmann A, Buitelaar J, Verhulst B, Fisher SE, Rai D, St Pourcain B. de Hoyos L, et al. Nat Commun. 2024 Feb 27;15(1):1770. doi: 10.1038/s41467-024-46128-8. Nat Commun. 2024. PMID: 38413609 Free PMC article. - Clinical characteristics of probands with obsessive-compulsive disorder from simplex and multiplex families.
Lima MO, Saraiva LC, Ramos VR, Oliveira MC, Costa DLC; Brazilian Research Consortium on Obsessive-Compulsive Spectrum Disorders; Fernandez TV, Crowley JJ, Storch EA, Shavitt RG, Miguel EC, Cappi C. Lima MO, et al. Psychiatry Res. 2024 Jan;331:115627. doi: 10.1016/j.psychres.2023.115627. Epub 2023 Nov 30. Psychiatry Res. 2024. PMID: 38113811
References
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. - PubMed
- Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev. 2012;22:229–237. - PubMed
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, McCague P, Dimiceli S, Pitts T, Nguyen L, Yang J, Harper C, Thorpe D, Vermeer S, Young H, Hebert J, Lin A, Ferguson J, Chiotti C, Wiese-Slater S, Rogers T, Salmon B, Nicholas P, Petersen PB, Pingree C, McMahon W, Wong DL, Cavalli-Sforza LL, Kraemer HC. et al.A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet. 1999;65:493–507. - PMC - PubMed
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimäki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K. et al.Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. - PMC - PubMed
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R. et al.Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials