Comparison of family history and SNPs for predicting risk of complex disease - PubMed (original) (raw)
Comparison of family history and SNPs for predicting risk of complex disease
Chuong B Do et al. PLoS Genet. 2012.
Abstract
The clinical utility of family history and genetic tests is generally well understood for simple Mendelian disorders and rare subforms of complex diseases that are directly attributable to highly penetrant genetic variants. However, little is presently known regarding the performance of these methods in situations where disease susceptibility depends on the cumulative contribution of multiple genetic factors of moderate or low penetrance. Using quantitative genetic theory, we develop a model for studying the predictive ability of family history and single nucleotide polymorphism (SNP)-based methods for assessing risk of polygenic disorders. We show that family history is most useful for highly common, heritable conditions (e.g., coronary artery disease), where it explains roughly 20%-30% of disease heritability, on par with the most successful SNP models based on associations discovered to date. In contrast, we find that for diseases of moderate or low frequency (e.g., Crohn disease) family history accounts for less than 4% of disease heritability, substantially lagging behind SNPs in almost all cases. These results indicate that, for a broad range of diseases, already identified SNP associations may be better predictors of risk than their family history-based counterparts, despite the large fraction of missing heritability that remains to be explained. Our model illustrates the difficulty of using either family history or SNPs for standalone disease prediction. On the other hand, we show that, unlike family history, SNP-based tests can reveal extreme likelihood ratios for a relatively large percentage of individuals, thus providing potentially valuable adjunctive evidence in a differential diagnosis.
Conflict of interest statement
CBD, DAH, UF, and NE are employed by 23andMe and own either stock or stock options in the company. PLOS co-founder Michael B. Eisen is a member of the 23andMe Scientific Advisory Board and also holds stock options in the company.
Figures
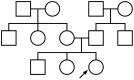
Figure 1. Test pedigree.
The test pedigree used throughout this paper, consisting of an extended family with multiple aunts and uncles. An arrow designates one particular individual as the “index individual” (or consultand) whose disease risk we wish to predict.
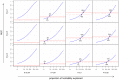
Figure 2. Area under the curve (AUC) plots.
Each cell of the  grid corresponds to a different combination of disease characteristics: rows correspond to differing heritabilities (
grid corresponds to a different combination of disease characteristics: rows correspond to differing heritabilities ( ) and columns correspond to differing frequencies (
) and columns correspond to differing frequencies ( ). Within each cell, the subplots compare the AUC when using a complete family history model that accounts for the disease status of every individual in the pedigree (solid red line), a restricted family history model that only considers the number (0, 1, or
). Within each cell, the subplots compare the AUC when using a complete family history model that accounts for the disease status of every individual in the pedigree (solid red line), a restricted family history model that only considers the number (0, 1, or  1) of affected first-degree relatives of the index individual (dashed red line), or genetic risk factors for the index individual only (blue line). For each subplot, the horizontal axis indicates the proportion (
1) of affected first-degree relatives of the index individual (dashed red line), or genetic risk factors for the index individual only (blue line). For each subplot, the horizontal axis indicates the proportion ( ) of heritability explained by known SNP associations, and the vertical axis indicates the AUC. Arrows indicate points of equivalence—values of
) of heritability explained by known SNP associations, and the vertical axis indicates the AUC. Arrows indicate points of equivalence—values of  and
and  for which family history and SNP-based methods give the same AUC.
for which family history and SNP-based methods give the same AUC.
Figure 3. Proportion of heritability explained.
Subpanels (A) and (B) contain contour plots showing the proportion of heritability explained ( ) by a complete family history model and a restricted family history model, respectively. Horizontal and vertical axes correspond to varying disease frequency (
) by a complete family history model and a restricted family history model, respectively. Horizontal and vertical axes correspond to varying disease frequency ( ) and heritability (
) and heritability ( ). Lines in each subplot depict the level curves of
). Lines in each subplot depict the level curves of  , i.e., the combinations of
, i.e., the combinations of  and
and  for which the proportion of heritability explained by family history is constant. SNP-based risk models for specific diseases are illustrated by circles (when the SNP-based model outperforms family history) and squares (when family history outperforms the SNP-based model). The circle or square for each SNP-based model has been colored to indicate the estimated proportion of heritability explained by SNPs, using the same color scheme as the contour plot (e.g., blue indicates 25–30% of heritability explained whereas red indicates
for which the proportion of heritability explained by family history is constant. SNP-based risk models for specific diseases are illustrated by circles (when the SNP-based model outperforms family history) and squares (when family history outperforms the SNP-based model). The circle or square for each SNP-based model has been colored to indicate the estimated proportion of heritability explained by SNPs, using the same color scheme as the contour plot (e.g., blue indicates 25–30% of heritability explained whereas red indicates  5% of heritability explained). Note that the performance of SNP-based models shown here reflects only currently known genetic factors for European populations and will change as more associations are discovered.
5% of heritability explained). Note that the performance of SNP-based models shown here reflects only currently known genetic factors for European populations and will change as more associations are discovered.
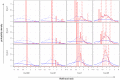
Figure 4. Distribution of likelihood ratios (LRs).
The subplots show density histograms for the distribution of LRs achieved by complete (solid red line) and restricted (dotted red line) family history models, and probability density functions for the distribution of LRs achieved by genetic factors accounting for either 10% (dotted blue line), 30% (dashed blue line), or 100% (solid blue line) of the heritability of the disease. As a technical aside, since all plots are shown on a logarithmic scale, the density histograms and density functions shown here were derived for  rather than
rather than  itself (see Text S1).
itself (see Text S1).
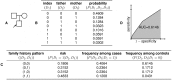
Figure 5. Worked example.
(A) In the family structure shown, the shaded box represents the index individual, whose risk of developing the disease we wish to predict. (B) There are  possible combinations of disease status for the individuals in the family. Using the liability threshold model, we compute the probability of each combination; in this example, we assume
possible combinations of disease status for the individuals in the family. Using the liability threshold model, we compute the probability of each combination; in this example, we assume  and
and  . (C) From the joint distribution, we can then compute the disease risk of the index individual for any given family history pattern, as well as the likelihood of particular family history patterns among cases and controls. (D) These quantities then allow us to construct the receiver operating characteristic (ROC) curve for a complete family history-based classifier, from which sensitivity, specificity, PPV, NPV, and AUC can be computed.
. (C) From the joint distribution, we can then compute the disease risk of the index individual for any given family history pattern, as well as the likelihood of particular family history patterns among cases and controls. (D) These quantities then allow us to construct the receiver operating characteristic (ROC) curve for a complete family history-based classifier, from which sensitivity, specificity, PPV, NPV, and AUC can be computed.
Similar articles
- Assessing thyroid cancer risk using polygenic risk scores.
Liyanarachchi S, Gudmundsson J, Ferkingstad E, He H, Jonasson JG, Tragante V, Asselbergs FW, Xu L, Kiemeney LA, Netea-Maier RT, Mayordomo JI, Plantinga TS, Hjartarson H, Hrafnkelsson J, Sturgis EM, Brock P, Nabhan F, Thorleifsson G, Ringel MD, Stefansson K, de la Chapelle A. Liyanarachchi S, et al. Proc Natl Acad Sci U S A. 2020 Mar 17;117(11):5997-6002. doi: 10.1073/pnas.1919976117. Epub 2020 Mar 4. Proc Natl Acad Sci U S A. 2020. PMID: 32132206 Free PMC article. - Prediction of lung cancer risk in a Chinese population using a multifactorial genetic model.
Li H, Yang L, Zhao X, Wang J, Qian J, Chen H, Fan W, Liu H, Jin L, Wang W, Lu D. Li H, et al. BMC Med Genet. 2012 Dec 10;13:118. doi: 10.1186/1471-2350-13-118. BMC Med Genet. 2012. PMID: 23228068 Free PMC article. - SNP-based heritability and genetic architecture of tarsal osteochondrosis in North American Standardbred horses.
McCoy AM, Norton EM, Kemper AM, Beeson SK, Mickelson JR, McCue ME. McCoy AM, et al. Anim Genet. 2019 Feb;50(1):78-81. doi: 10.1111/age.12738. Epub 2018 Oct 24. Anim Genet. 2019. PMID: 30353927 - Clinical utility of the polygenic LDL-C SNP score in familial hypercholesterolemia.
Futema M, Bourbon M, Williams M, Humphries SE. Futema M, et al. Atherosclerosis. 2018 Oct;277:457-463. doi: 10.1016/j.atherosclerosis.2018.06.006. Atherosclerosis. 2018. PMID: 30270085 Review. - Single Marker Family-Based Association Analysis Not Conditional on Parental Information.
Namkung J, Won S. Namkung J, et al. Methods Mol Biol. 2017;1666:409-439. doi: 10.1007/978-1-4939-7274-6_20. Methods Mol Biol. 2017. PMID: 28980257 Review.
Cited by
- Preimplantation Genetic Testing for Polygenic Disease Relative Risk Reduction: Evaluation of Genomic Index Performance in 11,883 Adult Sibling Pairs.
Treff NR, Eccles J, Marin D, Messick E, Lello L, Gerber J, Xu J, Tellier LCAM. Treff NR, et al. Genes (Basel). 2020 Jun 12;11(6):648. doi: 10.3390/genes11060648. Genes (Basel). 2020. PMID: 32545548 Free PMC article. - Risk Stratification and Clinical Utility of Polygenic Risk Scores in Ophthalmology.
Qassim A, Souzeau E, Hollitt G, Hassall MM, Siggs OM, Craig JE. Qassim A, et al. Transl Vis Sci Technol. 2021 May 3;10(6):14. doi: 10.1167/tvst.10.6.14. Transl Vis Sci Technol. 2021. PMID: 34111261 Free PMC article. - Endophenotypes for Age-Related Macular Degeneration: Extending Our Reach into the Preclinical Stages of Disease.
Gorin MB, Weeks DE, Baron RV, Conley YP, Ortube MC, Nusinowitz S. Gorin MB, et al. J Clin Med. 2014 Nov 28;3(4):1335-56. doi: 10.3390/jcm3041335. J Clin Med. 2014. PMID: 25568804 Free PMC article. - The SCRIPT trial: study protocol for a randomised controlled trial of a polygenic risk score to tailor colorectal cancer screening in primary care.
Saya S, Boyd L, Chondros P, McNamara M, King M, Milton S, Lourenco RA, Clark M, Fishman G, Marker J, Ostroff C, Allman R, Walter FM, Buchanan D, Winship I, McIntosh J, Macrae F, Jenkins M, Emery J. Saya S, et al. Trials. 2022 Sep 27;23(1):810. doi: 10.1186/s13063-022-06734-7. Trials. 2022. PMID: 36163034 Free PMC article. - Exposure-associated DNA methylation among people exposed to multiple industrial pollutants.
Chen CS, Yuan TH, Lu TP, Lee HY, Chen YH, Lai LC, Tsai MH, Chuang EY, Chan CC. Chen CS, et al. Clin Epigenetics. 2024 Aug 20;16(1):111. doi: 10.1186/s13148-024-01705-y. Clin Epigenetics. 2024. PMID: 39164771 Free PMC article.
References
- Maher B (2008) Personal genomes: The case of the missing heritability. Nature 456: 18–21. - PubMed
- So HC, Gui AH, Cherny SS, Sham PC (2011) Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol 35: 310–317. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources