Generation and characterization of a novel Cyp2a(4/5)bgs-null mouse model - PubMed (original) (raw)
Generation and characterization of a novel Cyp2a(4/5)bgs-null mouse model
Yuan Wei et al. Drug Metab Dispos. 2013 Jan.
Abstract
Knockout mouse models targeting various cytochrome P450 (P450 or CYP) genes are valuable for determining P450's biologic functions, including roles in drug metabolism and chemical toxicity. In this study, a novel Cyp2a(4/5)bgs-null mouse model was generated, in which a 1.2-megabase pair genomic fragment containing nine Cyp genes in mouse chromosome 7 (including, sequentially, Cyp2a5, 2g1, 2b19, 2b23, 2a4, 2b9, 2b13, 2b10, and 2s1) are deleted, through Cre-mediated recombination in vivo. The resultant mouse strain was viable and fertile, without any developmental deficits or morphologic abnormalities. Deletion of the constitutive genes in the cluster was confirmed by polymerase chain reaction analysis of the genes and the mRNAs in tissues known to express each gene. The loss of this gene cluster led to significant decreases in microsomal activities toward testosterone hydroxylation in various tissues examined, including olfactory mucosa (OM), lung, liver, and brain. In addition, systemic clearance of pentobarbital was decreased in Cyp2a(4/5)bgs-null mice, as indicated by >60% increases in pentobarbital-induced sleeping time, compared with wild-type (WT) mice. This novel Cyp2a(4/5)bgs-null mouse model will be valuable for in vivo studies of drug metabolism and chemical toxicities in various tissues, including the liver, lung, brain, intestine, kidney, skin, and OM, where one or more of the targeted Cyp genes are known to be expressed in WT mice. The model will also be valuable for preparation of humanized mice that express human CYP2A6, CYP2A13, CYP2B6, or CYP2S1, and as a knockout mouse model for five non-P450 genes (Vmn1r184, Nalp9c, Nalp4a, Nalp9a, and Vmn1r185) that were also deleted.
Figures
Fig. 1.
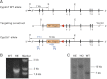
Targeted disruption of the mouse Cyp2s1 gene. (A) targeting strategy for Cyp2s1. The Cyp2s1 gene sequence is indicated by a thick line, whereas sequences from cloning vectors are shown in thin lines. Cyp2s1 exons are indicated as solid boxes. The positions of selected restriction sites (E, EcoR V; S, Spe I) are indicated. PCR primers used for genotyping (F1/R1) are shown as small thin arrows. DNA probes used for Southern blot analysis, P1 (external probe, 1.3-kb in size) and P2 (internal probe, 1.3 kb in size), are shown as blue bars. The diagnostic EcoR V fragments detected by probe P1 in WT allele (7 kb) and in targeted (Cyp2s1−) allele (10 kb), as well as the positions of PCR primers used for detecting Cyp2s1− allele, are indicated. (B) PCR analysis of genomic DNA from WT and Cyp2s1+/− mice. The expected 1.9-kb product was detected in the heterozygotes. Selected bands of a 1-kb DNA size marker are shown. (C) Southern blot analysis of genomic DNA with the external probe P1. Expected EcoR V fragments were detected in heterozygous (HE), homozygous (HO), and WT mice.
Fig. 2.
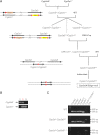
Generation of the _Cyp2a(4/5)bgs_-null mouse model. (A) breeding strategy. (B) identification of the #28-2 pup as harboring the Cyp2s1−/Cyp2a5− (double-knockout) allele. PCR detection of the Cyp2s1− and Cyp2a5− alleles was performed as described in methods. Representative Cyp2s1+/Cyp2a5- (lane 1) and Cyp2s1−/Cyp2a5+ (lane 3) samples, and the sample for the #28-2 pup (lane 2), are shown. (C) characterization of the _Cyp2a(4/5)bgs_-null allele. Genomic DNA from WT, Cyp2s1-Cyp2a5Δ/+, and Cyp2s1-Cyp2a5Δ/Δ mice was analyzed, using PCR primers for Cyp2a4, Cyp2a5, Cyp2g1, Cyp2b10, Cyp2s1, Cyp2s1-Cyp2a5Δ, and Cre. Selected bands of a 100-bp DNA size marker are shown.
Fig. 3.
Confirmation of gene cluster deletion in the _Cyp2a(4/5)bgs_-null (Cyp2s1-Cyp2a5Δ/Δ) mice. (A) strategy of Southern blot analysis. DNA probes P1 and P2 are shown as blue bars. Genomic DNA was digested by Apa I. Probe P1 would detect a 9.6-kb fragment from Cyp2a5 WT allele, whereas probe P2 would detect a 3.1-kb fragment from Cyp2s1 WT allele; both probes would detect a 6.6-kb fragment from the Cyp2s1-Cyp2a5Δ allele. (B) Southern blot analysis of WT and Cyp2s1-Cyp2a5Δ/Δ (HO) mice. Ten micrograms of genomic DNA were used for each lane. (C) RNA-PCR analysis of CYP2A4/5, CYP2B9/10/19, CYP2G1, and CYP2S1 expression in liver, lung, and OM of WT and _Cyp2a(4/5)bgs_-null mice. Tissues from 2-month-old (one male and one female) mice were pooled for total RNA preparation. PCR products were analyzed on a 1.5% agarose gel, and visualized by staining with ethidium bromide. The positions of selected fragments of a 100-bp DNA marker are indicated.
Fig. 4.
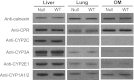
Immunoblot analysis of the expression of selected P450 and CPR. Microsomes were prepared from pooled liver, lung, or OM of WT and _Cyp2a(4/5)bgs_-null (Null) male mice (five per group, 2 months old). Microsomal proteins from the liver (5 _µ_g per lane), lung, and OM (10 _µ_g per lane) were analyzed for the expression of selected P450 or CPR proteins. The expression of calnexin was analyzed as a loading control. The antibodies used are described in Materials and Methods. Densitometric analysis (not shown) for each panel indicated that the maximal difference in band intensity between samples from WT and Null mice was less than 20%. Similar results were obtained in female mice (data not shown).
Similar articles
- Impact of nicotine metabolism on nicotine's pharmacological effects and behavioral responses: insights from a Cyp2a(4/5)bgs-null mouse.
Li L, Jia K, Zhou X, McCallum SE, Hough LB, Ding X. Li L, et al. J Pharmacol Exp Ther. 2013 Dec;347(3):746-54. doi: 10.1124/jpet.113.208256. Epub 2013 Sep 17. J Pharmacol Exp Ther. 2013. PMID: 24045421 Free PMC article. - Role of CYP2A5 in the clearance of nicotine and cotinine: insights from studies on a Cyp2a5-null mouse model.
Zhou X, Zhuo X, Xie F, Kluetzman K, Shu YZ, Humphreys WG, Ding X. Zhou X, et al. J Pharmacol Exp Ther. 2010 Feb;332(2):578-87. doi: 10.1124/jpet.109.162610. Epub 2009 Nov 18. J Pharmacol Exp Ther. 2010. PMID: 19923441 Free PMC article. - Generation and characterization of a CYP2A13/2B6/2F1-transgenic mouse model.
Wei Y, Wu H, Li L, Liu Z, Zhou X, Zhang QY, Weng Y, D'Agostino J, Ling G, Zhang X, Kluetzman K, Yao Y, Ding X. Wei Y, et al. Drug Metab Dispos. 2012 Jun;40(6):1144-50. doi: 10.1124/dmd.112.044826. Epub 2012 Mar 7. Drug Metab Dispos. 2012. PMID: 22397853 Free PMC article. - A CYP2B6-humanized mouse model and its potential applications.
Li L, Zhang QY, Ding X. Li L, et al. Drug Metab Pharmacokinet. 2018 Feb;33(1):2-8. doi: 10.1016/j.dmpk.2018.01.001. Epub 2018 Jan 11. Drug Metab Pharmacokinet. 2018. PMID: 29402634 Review. - Cytochrome P450 knockout mice: new toxicological models.
McKinnon RA, Nebert DW. McKinnon RA, et al. Clin Exp Pharmacol Physiol. 1998 Oct;25(10):783-7. doi: 10.1111/j.1440-1681.1998.tb02153.x. Clin Exp Pharmacol Physiol. 1998. PMID: 9784916 Review.
Cited by
- Suppression of pulmonary CYP2A13 expression by carcinogen-induced lung tumorigenesis in a CYP2A13-humanized mouse model.
Liu Z, Megaraj V, Li L, Sell S, Hu J, Ding X. Liu Z, et al. Drug Metab Dispos. 2015 May;43(5):698-702. doi: 10.1124/dmd.115.063305. Epub 2015 Feb 20. Drug Metab Dispos. 2015. PMID: 25710941 Free PMC article. - Maternally contributed Nlrp9b expressed in human and mouse ovarian follicles contributes to early murine preimplantation development.
Amoushahi M, Steffensen LL, Galieva A, Agger J, Heuck A, Siupka P, Ernst E, Nielsen MS, Sunde L, Lykke-Hartmann K. Amoushahi M, et al. J Assist Reprod Genet. 2020 Jun;37(6):1355-1365. doi: 10.1007/s10815-020-01767-w. Epub 2020 May 12. J Assist Reprod Genet. 2020. PMID: 32399794 Free PMC article. - A transcriptomic approach to elucidate the physiological significance of human cytochrome P450 2S1 in bronchial epithelial cells.
Madanayake TW, Lindquist IE, Devitt NP, Mudge J, Rowland AM. Madanayake TW, et al. BMC Genomics. 2013 Nov 26;14:833. doi: 10.1186/1471-2164-14-833. BMC Genomics. 2013. PMID: 24279958 Free PMC article. - NLRP9B protein is dispensable for oocyte maturation and early embryonic development in the mouse.
Peng H, Lin X, Liu F, Wang C, Zhang W. Peng H, et al. J Reprod Dev. 2015;61(6):559-64. doi: 10.1262/jrd.2015-050. Epub 2015 Sep 28. J Reprod Dev. 2015. PMID: 26411641 Free PMC article. - The Role of Nicotine Metabolic Rate on Nicotine Dependence and Rewarding: Nicotine Metabolism in Chinese Male Smokers and Male Mice.
Liu M, Wang H, Fu Y, Li X, Wu P, Liu G, Wang R, Zhang Y, Chen H, Hou H, Hu Q. Liu M, et al. Mol Neurobiol. 2024 Oct;61(10):7692-7706. doi: 10.1007/s12035-024-04040-8. Epub 2024 Mar 1. Mol Neurobiol. 2024. PMID: 38427211
References
Publication types
MeSH terms
Substances
Grants and funding
- ES001337/ES/NIEHS NIH HHS/United States
- CA092596/CA/NCI NIH HHS/United States
- R01 CA092596/CA/NCI NIH HHS/United States
- ES007462/ES/NIEHS NIH HHS/United States
- R01 ES007462/ES/NIEHS NIH HHS/United States
- R01 GM082978/GM/NIGMS NIH HHS/United States
- GM082978/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases