Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia - PubMed (original) (raw)
Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia
Malek Faham et al. Blood. 2012.
Abstract
The persistence of minimal residual disease (MRD) during therapy is the strongest adverse prognostic factor in acute lymphoblastic leukemia (ALL). We developed a high-throughput sequencing method that universally amplifies antigen-receptor gene segments and identifies all clonal gene rearrangements (ie, leukemia-specific sequences) at diagnosis, allowing monitoring of disease progression and clonal evolution during therapy. In the present study, the assay specifically detected 1 leukemic cell among greater than 1 million leukocytes in spike-in experiments. We compared this method with the gold-standard MRD assays multiparameter flow cytometry and allele-specific oligonucleotide polymerase chain reaction (ASO-PCR) using diagnostic and follow-up samples from 106 patients with ALL. Sequencing detected MRD in all 28 samples shown to be positive by flow cytometry and in 35 of the 36 shown to be positive by ASO-PCR and revealed MRD in 10 and 3 additional samples that were negative by flow cytometry and ASO-PCR, respectively. We conclude that this new method allows monitoring of treatment response in ALL and other lymphoid malignancies with great sensitivity and precision. The www.clinicaltrials.gov identifier number for the Total XV study is NCT00137111.
Figures
Figure 1
Overview of the LymphoSIGHT method. (A) Schematic of the PCR primer strategy and sequencing assay. (B) Schematic of the MRD quantitation scheme.
Figure 2
Technical performance of immune repertoire sequencing assay. Diagnostic samples from 12 ALL patients containing 13 leukemic IgH clonotypes were used for technical performance studies. Serial dilutions of leukemic cells in normal PBMCs, ranging from < 1 in 1 million to > 1 in 1000 tumor cells, were prepared and analyzed in duplicate. The 7 duplicated dilutions were then subjected to amplification and sequencing in 2 replicate experiments. The expected and observed frequencies were compared on a logarithmic scale. Replicate 1 is represented by circles and replicate 2 is represented by crosses in all panels. Panel G shows the results from 2 cancer clones that were present in this tumor.
Figure 3
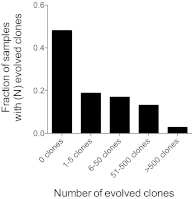
Clonal evolution mechanisms at the IgH gene locus. Diagnostic samples containing a clonal gene rearrangement, as determined using the VDJ (n = 106) assay were categorized into 5 groups based on the number of evolved clones present in the diagnostic sample. The fraction of all samples having a given number of evolved clones is shown.
Figure 4
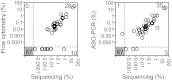
Comparison of MRD results obtained by sequencing, flow cytometry, and ASO-PCR. MRD results obtained using the sequencing method were compared with flow cytometry results for 105 ALL patients (left) and with ASO-PCR results for 106 ALL patients (right). In each panel, the numbers of concordant measurements are shown in the lower left and upper right and the number of discordant measurements are shown in the upper left and lower right.
Figure 5
Schematic diagram of sequencing analysis and work flow for prospective sample collection.
Similar articles
- An update on PCR use for minimal residual disease monitoring in acute lymphoblastic leukemia.
Nunes V, Cazzaniga G, Biondi A. Nunes V, et al. Expert Rev Mol Diagn. 2017 Nov;17(11):953-963. doi: 10.1080/14737159.2017.1377073. Epub 2017 Sep 21. Expert Rev Mol Diagn. 2017. PMID: 28891364 Review. - How I investigate minimal residual disease in acute lymphoblastic leukemia.
Correia RP, Bento LC, de Sousa FA, Barroso RS, Campregher PV, Bacal NS. Correia RP, et al. Int J Lab Hematol. 2021 Jun;43(3):354-363. doi: 10.1111/ijlh.13463. Epub 2021 Jan 10. Int J Lab Hematol. 2021. PMID: 33423385 Review. - Minimal residual disease (MRD) detection using rearrangement of immunoglobulin/T cell receptor genes in adult patients with acute lymphoblastic leukemia (ALL).
Shahkarami S, Mehrasa R, Younesian S, Yaghmaie M, Chahardouli B, Vaezi M, Rezaei N, Nikbakht M, Alimoghaddam K, Ghavamzadeh A, Tavakkoly-Bazzaz J, Ghaffari SH. Shahkarami S, et al. Ann Hematol. 2018 Apr;97(4):585-595. doi: 10.1007/s00277-018-3230-z. Epub 2018 Feb 1. Ann Hematol. 2018. PMID: 29392424 - Primers and protocols for standardized detection of minimal residual disease in acute lymphoblastic leukemia using immunoglobulin and T cell receptor gene rearrangements and TAL1 deletions as PCR targets: report of the BIOMED-1 CONCERTED ACTION: investigation of minimal residual disease in acute leukemia.
Pongers-Willemse MJ, Seriu T, Stolz F, d'Aniello E, Gameiro P, Pisa P, Gonzalez M, Bartram CR, Panzer-Grümayer ER, Biondi A, San Miguel JF, van Dongen JJ. Pongers-Willemse MJ, et al. Leukemia. 1999 Jan;13(1):110-8. doi: 10.1038/sj.leu.2401245. Leukemia. 1999. PMID: 10049045
Cited by
- Certainty in uncertainty: Determining the rate and reasons for reclassification of variants of uncertain significance in haematological malignancies.
Enjeti AK, Walker N, Fahey O, Johnston E, Legge-Wilkinson H, Ramsurrun N, Sillar J, Lincz LF, Ziolkowski A, Mossman D. Enjeti AK, et al. EJHaem. 2024 Sep 12;5(5):957-963. doi: 10.1002/jha2.1002. eCollection 2024 Oct. EJHaem. 2024. PMID: 39415915 Free PMC article. - Monitoring measurable residual disease in paediatric acute lymphoblastic leukaemia using immunoglobulin gene clonality based on next-generation sequencing.
Ahn WK, Yu K, Kim H, Lee ST, Choi JR, Han JW, Lyu CJ, Hahn S, Shin S. Ahn WK, et al. Cancer Cell Int. 2024 Jun 25;24(1):218. doi: 10.1186/s12935-024-03404-3. Cancer Cell Int. 2024. PMID: 38918782 Free PMC article. - The Evolving Landscape of Flowcytometric Minimal Residual Disease Monitoring in B-Cell Precursor Acute Lymphoblastic Leukemia.
Verbeek MWC, van der Velden VHJ. Verbeek MWC, et al. Int J Mol Sci. 2024 Apr 30;25(9):4881. doi: 10.3390/ijms25094881. Int J Mol Sci. 2024. PMID: 38732101 Free PMC article. Review. - Evaluation of next-generation sequencing versus next-generation flow cytometry for minimal-residual-disease detection in Chinese patients with multiple myeloma.
Zhou M, Chen Y, Gong Y, Zhu M, Cen J, Pan J, Yan L, Shang J, Jin S, Shi X, Yao W, Yan S, Wu D, Chen S, Fu C, Yao L. Zhou M, et al. Discov Oncol. 2024 Mar 19;15(1):78. doi: 10.1007/s12672-024-00938-w. Discov Oncol. 2024. PMID: 38502423 Free PMC article. - Relationship between subtype-specific minimal residual disease level and long-term prognosis in children with acute lymphoblastic leukemia.
Huang XT, Wang CJ, Gao C, Xue TL, Zhao ZJ, Wang TY, Wu MY, Cui L, Zhang RD, Li ZG. Huang XT, et al. Ann Hematol. 2024 Sep;103(9):3657-3665. doi: 10.1007/s00277-024-05687-y. Epub 2024 Mar 18. Ann Hematol. 2024. PMID: 38494553
References
- Gökbuget N, Hoelzer D. Treatment of adult acute lymphoblastic leukemia. Semin Hematol. 2009;46(1):64–75. - PubMed
- Brüggemann M, Schrauder A, Raff T, et al. Standardized MRD quantification in European ALL trials: proceedings of the Second International Symposium on MRD assessment in Kiel, Germany, 18-20 September 2008. Leukemia. 2010;24(3):521–535. - PubMed
- Campana D. Minimal residual disease in acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program. 2010;2010:7–12. - PubMed
- van der Velden VH, Cazzaniga G, Schrauder A, et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia. 2007;21(4):604–611. - PubMed
Publication types
MeSH terms
Grants and funding
- CA21765/CA/NCI NIH HHS/United States
- R01 CA060419/CA/NCI NIH HHS/United States
- CA60419/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- U01 CA060419/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources