Inhibition of reverse transcriptase activity increases stability of the HIV-1 core - PubMed (original) (raw)
Inhibition of reverse transcriptase activity increases stability of the HIV-1 core
Yang Yang et al. J Virol. 2013 Jan.
Abstract
Previous studies showed that HIV-1 reverse transcription occurs during or before uncoating, linking mechanistically reverse transcription with uncoating. Here we show that inhibition of reverse transcriptase (RT) during HIV-1 infection by pharmacologic or genetic means increased the stability of the HIV-1 core during infection. Interestingly, HIV-1 particles with increased core stability were resistant to the core-destabilizing effects of rhesus TRIM5α (TRIM5α(rh)). Collectively, this work implies that the surface of the HIV-1 core is dynamic and changes upon the ongoing processes within the core.
Figures
Fig 1
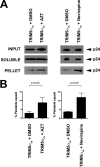
Effects of reverse transcriptase inhibitors on HIV-1 core stability. (A) Cf2Th cells transduced with the empty vector LPCX were challenged with increasing amounts of HIV-1 GFP-reporter virus (800 pg/ml of p24) in the presence of AZT. As a control, LPCX-transduced cells were challenged in the presence of DMSO, the solvent used to resuspend the RT inhibitors. GFP-positive cells were quantified by flow cytometry. (B) LPCX-transduced Cf2Th cells in the presence of AZT or DMSO were challenged with similar amounts of HIV-1 GFP-reporter, and the amount of soluble versus particulate capsid was determined by the FOC assay. As control, we used the same amount of HIV-1 GFP-reporter to perform the FOC assay in Cf2Th cells stably expressing TRIM5αrh, which is expressed from the LPCX vector. Briefly, cells were incubated with HIV-1 GFP-reporter at 4°C for 30 min, washed, and returned to 37°C. Infection was allowed to proceed for 16 h. Cell extracts were fractionated on a sucrose cushion. Input, soluble, and pellet fractions were analyzed by Western blotting using antibodies against the HIV-1 p24 capsid protein. (C) The percentage of pelletable HIV-1 capsid was determined with respect to the amount of total input capsid. Similar results were obtained in three independent experiments, and standard deviations are shown. Statistical differences are given as P < 0.001 (two-way analysis of variance [ANOVA] followed by the Bonferroni posttest). (D) Cf2Th cells stably transduced with TRIM5αrh and selected in puromycin were analyzed for TRIM5αrh-hemagglutinin (HA) expression by Western blotting using anti-HA antibodies. The loading control was performed using anti-β-actin antibodies.
Fig 2
Effects of RT mutants on stability of the HIV-1 core. (A) Cf2Th cells transduced with the empty vector LPCX were challenged with the indicated HIV-1-GFP reporter viruses normalized by ELISA against p24 (800 pg/ml of p24). To study the effect of reverse transcription on core stability, we challenged cells with an HIV-1 strain bearing a mutation in the active site of the RT enzyme (HIV-1 RT D185N). Forty-eight hours postinfection, GFP-positive cells were quantified by flow cytometry. (B) Because the HIV-1 RT D185N mutant is not infectious, Gag-processing levels in viral supernatants were evaluated by Western blotting using antibodies against p24. (C) LPCX-transduced Cf2Th cells were challenged with similar amounts of the indicated HIV-1 GFP-reporter viruses, and the amount of soluble versus particulate capsid was determined by FOC assay. As a control, we used the same amount of wild-type HIV-1 GFP-reporter virus to perform the FOC assay in Cf2Th cells stably expressing TRIM5αrh, which is expressed from the LPCX vector. Briefly, cells were incubated with the indicated viruses at 4°C for 30 min, washed, and returned to 37°C. Infection was allowed to proceed for 16 h. Cell extracts were fractionated on a sucrose cushion. Input, soluble, and pellet fractions were analyzed by Western blotting using antibodies against the HIV-1 p24 capsid protein. (D) The percentage of pelletable HIV-1 capsid was determined with respect to the amount of total input capsid. Similar results were obtained in three independent experiments, and standard deviations are shown. Statistical differences are given as P < 0.01 (two-way ANOVA followed by the Bonferroni posttest).
Fig 3
The core-stabilizing HIV-1 RT mutant is resistant to the destabilizing effects of TRIM5αrh. (A) Cf2Th cells transduced with the empty vector LPCX or expressing TRIM5αrh were challenged with the indicated HIV-1 GFP-reporter viruses normalized by ELISA against p24. The amount of soluble versus particulate capsid was determined by FOC assay. Briefly, cells were incubated with the indicated viruses at 4°C for 30 min, washed, and returned to 37°C. Infection was allowed to proceed for 16 h. Cell extracts were fractionated on a sucrose cushion. Input, soluble, and pellet fractions were analyzed by Western blotting using antibodies against HIV-1 p24 capsid protein. (B) The percentage of pelletable HIV-1 capsid was determined with respect to input capsid. Similar results were obtained in three independent experiments, and standard deviations are shown. Statistical differences are given as P < 0.001 (two-way ANOVA followed by the Bonferroni posttest).
Fig 4
The use of AZT and nevirapine prevents the core-destabilizing effects of TRIM5αrh. (A) Cf2Th cells stably transduced with TRIM5αrh were challenged with the indicated HIV-1 GFP-reporter viruses, normalized by ELISA against p24, in the presence of AZT and nevirapine. The amount of soluble versus particulate capsid was determined by FOC assay. Briefly, cells were incubated with the indicated viruses at 4°C for 30 min, washed, and returned to 37°C. Infection was allowed to proceed for 16 h. Cell extracts were fractionated on a sucrose cushion. Input, soluble, and pellet fractions were analyzed by Western blotting using antibodies against HIV-1 p24 capsid protein. (B) The percentage of pelletable HIV-1 capsid was determined with respect to input capsid. Similar results were obtained in three independent experiments, and standard deviations are shown. Statistical differences are given as P < 0.05 (two-way ANOVA followed by the Bonferroni posttest).
Similar articles
- RING domain mutations uncouple TRIM5α restriction of HIV-1 from inhibition of reverse transcription and acceleration of uncoating.
Roa A, Hayashi F, Yang Y, Lienlaf M, Zhou J, Shi J, Watanabe S, Kigawa T, Yokoyama S, Aiken C, Diaz-Griffero F. Roa A, et al. J Virol. 2012 Feb;86(3):1717-27. doi: 10.1128/JVI.05811-11. Epub 2011 Nov 23. J Virol. 2012. PMID: 22114335 Free PMC article. - A mutant tat protein inhibits HIV-1 reverse transcription by targeting the reverse transcription complex.
Lin MH, Apolloni A, Cutillas V, Sivakumaran H, Martin S, Li D, Wei T, Wang R, Jin H, Spann K, Harrich D. Lin MH, et al. J Virol. 2015 May;89(9):4827-36. doi: 10.1128/JVI.03440-14. Epub 2015 Feb 11. J Virol. 2015. PMID: 25673710 Free PMC article. - Oxazole-Benzenesulfonamide Derivatives Inhibit HIV-1 Reverse Transcriptase Interaction with Cellular eEF1A and Reduce Viral Replication.
Rawle DJ, Li D, Wu Z, Wang L, Choong M, Lor M, Reid RC, Fairlie DP, Harris J, Tachedjian G, Poulsen SA, Harrich D. Rawle DJ, et al. J Virol. 2019 May 29;93(12):e00239-19. doi: 10.1128/JVI.00239-19. Print 2019 Jun 15. J Virol. 2019. PMID: 30918071 Free PMC article. - Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors.
Sluis-Cremer N, Tachedjian G. Sluis-Cremer N, et al. Virus Res. 2008 Jun;134(1-2):147-56. doi: 10.1016/j.virusres.2008.01.002. Epub 2008 Mar 26. Virus Res. 2008. PMID: 18372072 Free PMC article. Review. - [Impacts of HIV-1 resistance mutations associated with nucleoside reverse transcriptase inhibitors on viral fitness].
Zhou Q, Liao LJ, Huang HJ. Zhou Q, et al. Bing Du Xue Bao. 2012 May;28(3):291-6. Bing Du Xue Bao. 2012. PMID: 22764534 Review. Chinese.
Cited by
- Complementary Assays Reveal a Low Level of CA Associated with Viral Complexes in the Nuclei of HIV-1-Infected Cells.
Hulme AE, Kelley Z, Foley D, Hope TJ. Hulme AE, et al. J Virol. 2015 May;89(10):5350-61. doi: 10.1128/JVI.00476-15. Epub 2015 Mar 4. J Virol. 2015. PMID: 25741002 Free PMC article. - HIV-1 capsid uncoating initiates after the first strand transfer of reverse transcription.
Cosnefroy O, Murray PJ, Bishop KN. Cosnefroy O, et al. Retrovirology. 2016 Aug 22;13(1):58. doi: 10.1186/s12977-016-0292-7. Retrovirology. 2016. PMID: 27549239 Free PMC article. - Allosteric HIV-1 Integrase Inhibitors Lead to Premature Degradation of the Viral RNA Genome and Integrase in Target Cells.
Madison MK, Lawson DQ, Elliott J, Ozantürk AN, Koneru PC, Townsend D, Errando M, Kvaratskhelia M, Kutluay SB. Madison MK, et al. J Virol. 2017 Aug 10;91(17):e00821-17. doi: 10.1128/JVI.00821-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28615207 Free PMC article. - Distinct nucleic acid interaction properties of HIV-1 nucleocapsid protein precursor NCp15 explain reduced viral infectivity.
Wang W, Naiyer N, Mitra M, Li J, Williams MC, Rouzina I, Gorelick RJ, Wu Z, Musier-Forsyth K. Wang W, et al. Nucleic Acids Res. 2014 Jun;42(11):7145-59. doi: 10.1093/nar/gku335. Epub 2014 May 9. Nucleic Acids Res. 2014. PMID: 24813443 Free PMC article. - HIV-1 Capsid Stabilization Assay.
Fricke T, Diaz-Griffero F. Fricke T, et al. Methods Mol Biol. 2016;1354:39-47. doi: 10.1007/978-1-4939-3046-3_3. Methods Mol Biol. 2016. PMID: 26714703 Free PMC article.
References
- Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11:672–675 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources