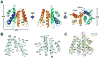Nucleic acid binding surface and dimer interface revealed by CRISPR-associated CasB protein structures - PubMed (original) (raw)
Nucleic acid binding surface and dimer interface revealed by CRISPR-associated CasB protein structures
Ki Hyun Nam et al. FEBS Lett. 2012.
Abstract
The CRISPR system is an adaptive RNA-based microbial immune system against invasive genetic elements. CasB is an essential protein component in Type I-E Cascade. Here, we characterize CasB proteins from three different organisms as non-specific nucleic acid binding proteins. The Thermobifida fusca CasB crystal structure reveals conserved positive surface charges, which we show are important for its nucleic acid binding function. EM docking reveals that CasB dimerization aligns individual nucleic acid binding surfaces into a curved, elongated binding surface inside Type I-E Cascade, consistent with the putative functions of CasB in ds-DNA recruitment and crRNA-DNA duplex formation steps.
Copyright © 2012 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures
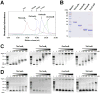
Fig. 1
CRISPR-associate CasB proteins bind nucleic acid nonspecifically. (A) Oligomeric state of CasB proteins as revealed by SEC. (B) SDS-PAGE of the purified CasB proteins. EMSA of CasB proteins binding to (C) dsDNA and (D) ssRNA. The purified CasB proteins were incubated with dsDNA (2.8 μM) or ssRNA (0.5 μM) at 25°C for 30 min.
Fig. 2
Crystal structure of TfuCasB2. (A) The α-helical bundle architecture in the crystal structure of TfuCasB2 (residues 11-196). (B) Dissection of TfuCasB2 in half to reveal that the hydrophobic core is stabilized by van der Waals contacts. (C) Structure superposition of TfuCasB2 (in cyan) and TthCasB (in yellow) reveals an identical core with different structural features in the peripheral regions (Cα r.m.s deviation of 2.2 Å).
Fig. 3
A conserved positive surface patch important for nucleic acid binding in TfuCasB2. (A) Electrostatic surface and (B) Surface conservation in the TfuCasB2. Residues are colored from magenta to cyan with descending order of conservation. (C) SDS-PAGE of the purified wild-type and mutant TfuCasB2 proteins. (D) EMSA for the binding of wild-type and mutant TfuCasB2 proteins to the dsDNA in Figure 1D. A 2-fold titration of CasB proteins from 3.125 μM to 50 μM were incubated with 1.1 μM ds-DNA at 25°C for 30 min. (E) Superimposition of TfuCasB2 (cyan) with TthCasB (yellow) showing that locations of the positively charged residues on the putative nucleic acid binding surface are highly conserved.
Fig. 4
Asymmetric CasB dimer formation from cryo-EM docking and crystal packing analysis. (A) Overall architecture of the E. coli Cascade complex (EMDB code 3514). CasA to E are colored in magenta, grey, yellow, cyan, and orange, respectively. The crRNA-ssDNA duplex is in green. An asymmetric CasB dimer is positioned along the helical spine composed of crRNA and six copies of CasC, connecting the head (CasA) and tail (CasE) of the E. coli Cascade. (B) Close-up view of two TfuCasB2 molecules docked into the E. coli CasB envelope. (C) Superimposition of the EM-docked TfuCasB2 dimer with a TthCasB dimer formed through crystal contacts. The dimer interface in (D) TfuCasB2 and (E) TthCasB involves hydrogen bonds and salt bridges, but residues involved are not conserved. (F) Close-up view of the cryo-EM density in E. coli Cascade (EMDB code 3515), showing that the CasB dimer makes multiple contacts (red-dotted circled) over a long distance on the crRNA/ss-DNA duplex (G) Electrostatistic surface representation of the EM-docked TthCasB dimer showing that the nucleic acid binding surface of each CasB monomer aligns into an extended, curved binding site upon dimer formation in Cascade.
Similar articles
- Structural insights into specific crRNA G-rich sequence binding by Meiothermus ruber Cse2.
Liu S, Yuan Z, Yuan YA. Liu S, et al. J Struct Biol. 2015 May;190(2):122-34. doi: 10.1016/j.jsb.2015.03.001. Epub 2015 Mar 16. J Struct Biol. 2015. PMID: 25791617 - Crystal structure of TTHA1264, a putative M16-family zinc peptidase from Thermus thermophilus HB8 that is homologous to the beta subunit of mitochondrial processing peptidase.
Ohtsuka J, Ichihara Y, Ebihara A, Nagata K, Tanokura M. Ohtsuka J, et al. Proteins. 2009 May 15;75(3):774-80. doi: 10.1002/prot.22365. Proteins. 2009. PMID: 19241474 No abstract available. - Mechanistic basis of plasmid-specific DNA binding of the F plasmid regulatory protein, TraM.
Peng Y, Lu J, Wong JJW, Edwards RA, Frost LS, Mark Glover JN. Peng Y, et al. J Mol Biol. 2014 Nov 11;426(22):3783-3795. doi: 10.1016/j.jmb.2014.09.018. Epub 2014 Oct 2. J Mol Biol. 2014. PMID: 25284757 - Bacterial replication initiator DnaA. Rules for DnaA binding and roles of DnaA in origin unwinding and helicase loading.
Messer W, Blaesing F, Jakimowicz D, Krause M, Majka J, Nardmann J, Schaper S, Seitz H, Speck C, Weigel C, Wegrzyn G, Welzeck M, Zakrzewska-Czerwinska J. Messer W, et al. Biochimie. 2001 Jan;83(1):5-12. doi: 10.1016/s0300-9084(00)01216-5. Biochimie. 2001. PMID: 11254968 Review. - Synthetic Life with Alternative Nucleic Acids as Genetic Materials.
Nie P, Bai Y, Mei H. Nie P, et al. Molecules. 2020 Jul 31;25(15):3483. doi: 10.3390/molecules25153483. Molecules. 2020. PMID: 32751873 Free PMC article. Review.
Cited by
- DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes.
Plagens A, Richter H, Charpentier E, Randau L. Plagens A, et al. FEMS Microbiol Rev. 2015 May;39(3):442-63. doi: 10.1093/femsre/fuv019. Epub 2015 Apr 30. FEMS Microbiol Rev. 2015. PMID: 25934119 Free PMC article. Review. - Exploiting CRISPR/Cas: interference mechanisms and applications.
Richter H, Randau L, Plagens A. Richter H, et al. Int J Mol Sci. 2013 Jul 12;14(7):14518-31. doi: 10.3390/ijms140714518. Int J Mol Sci. 2013. PMID: 23857052 Free PMC article. Review. - Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli.
Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, Brouns SJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. Jackson RN, et al. Science. 2014 Sep 19;345(6203):1473-9. doi: 10.1126/science.1256328. Epub 2014 Aug 7. Science. 2014. PMID: 25103409 Free PMC article. - DNA binding properties of the small cascade subunit Csa5.
Daume M, Plagens A, Randau L. Daume M, et al. PLoS One. 2014 Aug 22;9(8):e105716. doi: 10.1371/journal.pone.0105716. eCollection 2014. PLoS One. 2014. PMID: 25148031 Free PMC article. - Role of nucleotide identity in effective CRISPR target escape mutations.
Künne T, Zhu Y, da Silva F, Konstantinides N, McKenzie RE, Jackson RN, Brouns SJ. Künne T, et al. Nucleic Acids Res. 2018 Nov 2;46(19):10395-10404. doi: 10.1093/nar/gky687. Nucleic Acids Res. 2018. PMID: 30107450 Free PMC article.
References
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM086766/GM/NIGMS NIH HHS/United States
- P41 RR001646/RR/NCRR NIH HHS/United States
- GM-086766/GM/NIGMS NIH HHS/United States
- P41 RR015301/RR/NCRR NIH HHS/United States
- R01 GM102543/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases